Biomaterials Translational ›› 2023, Vol. 4 ›› Issue (3): 180-195.doi: 10.12336/biomatertransl.2023.03.006
• RESEARCH ARTICLE • Previous Articles
Rui Li1,2, Shuai Qiu3, Weihong Yang2,4, Zilong Rao1, Jiaxin Chen1, Yuexiong Yang4, Qingtang Zhu3,*( ), Xiaolin Liu3, Ying Bai1,*(
), Xiaolin Liu3, Ying Bai1,*( ), Daping Quan1,*(
), Daping Quan1,*( )
)
Received:2022-11-17
Revised:2022-12-06
Accepted:2023-03-08
Online:2023-09-28
Published:2023-09-28
Contact:
*Qingtang Zhu, Li, R.; Qiu, S.; Yang, W.; Rao, Z.; Chen, J.; Yang, Y.; Zhu, Q.; Liu, X.; Bai, Y.; Quan. D. A comparative study of human and porcine-derived decellularised nerve matrices. Biomater Transl. 2023, 4(3), 180-195.
| Gene | Primer sequence | Product size (bp) |
|---|---|---|
| MAP–2 | Forward: 5'–CTT CAC GCA CAC CAG GCA CTC–3' | 102 |
| Reverse: 5'–CCT TCT TCT CAC TCG GCA CCA AG–3' | ||
| GAP43 | Forward: 5'–TCC ACT GAT AAC TCG CCG TCC TC–3' | 94 |
| Reverse: 5'–CAG CAG CAG TGA CAG CAG CAG–3' | ||
| MBP | Forward: 5'–CGA GGA CGG AGA TGA GGA GTA GTC–3' | 197 |
| Reverse: 5'–CAG CTC AGC GAC GCA GAG TG–3' | ||
| MPZ | Forward: 5'–TGG TGC TGT TGC TGC TGC TG–3' | 185 |
| Reverse: 5'–GGT GCT TCT GCT GTG GTC CAG–3' | ||
| GFAP | Forward: 5'–GCT GCG GCT CGA TCA ACT CAC–3' | 169 |
| Reverse: 5'–GGT GGC TTC ATC TGC TTC CTG TC–3' | ||
| S100β | Forward: 5'–ACA ATG ATG GAG ACG GCG AAT GTG–3' | 80 |
| Reverse: 5'–GAA CTC GTG GCA GGC AGT AGT AAC–3' | ||
| β–Actin | Forward: 5'–GCA AGT GCT TCT AGG CGG ACT G–3' | 195 |
| Reverse: 5'–CTG CTG TCA CCT TCA CCG TTC C–3' |
Table 1. Primer sequences used for quantitative polymerase chain reaction
| Gene | Primer sequence | Product size (bp) |
|---|---|---|
| MAP–2 | Forward: 5'–CTT CAC GCA CAC CAG GCA CTC–3' | 102 |
| Reverse: 5'–CCT TCT TCT CAC TCG GCA CCA AG–3' | ||
| GAP43 | Forward: 5'–TCC ACT GAT AAC TCG CCG TCC TC–3' | 94 |
| Reverse: 5'–CAG CAG CAG TGA CAG CAG CAG–3' | ||
| MBP | Forward: 5'–CGA GGA CGG AGA TGA GGA GTA GTC–3' | 197 |
| Reverse: 5'–CAG CTC AGC GAC GCA GAG TG–3' | ||
| MPZ | Forward: 5'–TGG TGC TGT TGC TGC TGC TG–3' | 185 |
| Reverse: 5'–GGT GCT TCT GCT GTG GTC CAG–3' | ||
| GFAP | Forward: 5'–GCT GCG GCT CGA TCA ACT CAC–3' | 169 |
| Reverse: 5'–GGT GGC TTC ATC TGC TTC CTG TC–3' | ||
| S100β | Forward: 5'–ACA ATG ATG GAG ACG GCG AAT GTG–3' | 80 |
| Reverse: 5'–GAA CTC GTG GCA GGC AGT AGT AAC–3' | ||
| β–Actin | Forward: 5'–GCA AGT GCT TCT AGG CGG ACT G–3' | 195 |
| Reverse: 5'–CTG CTG TCA CCT TCA CCG TTC C–3' |
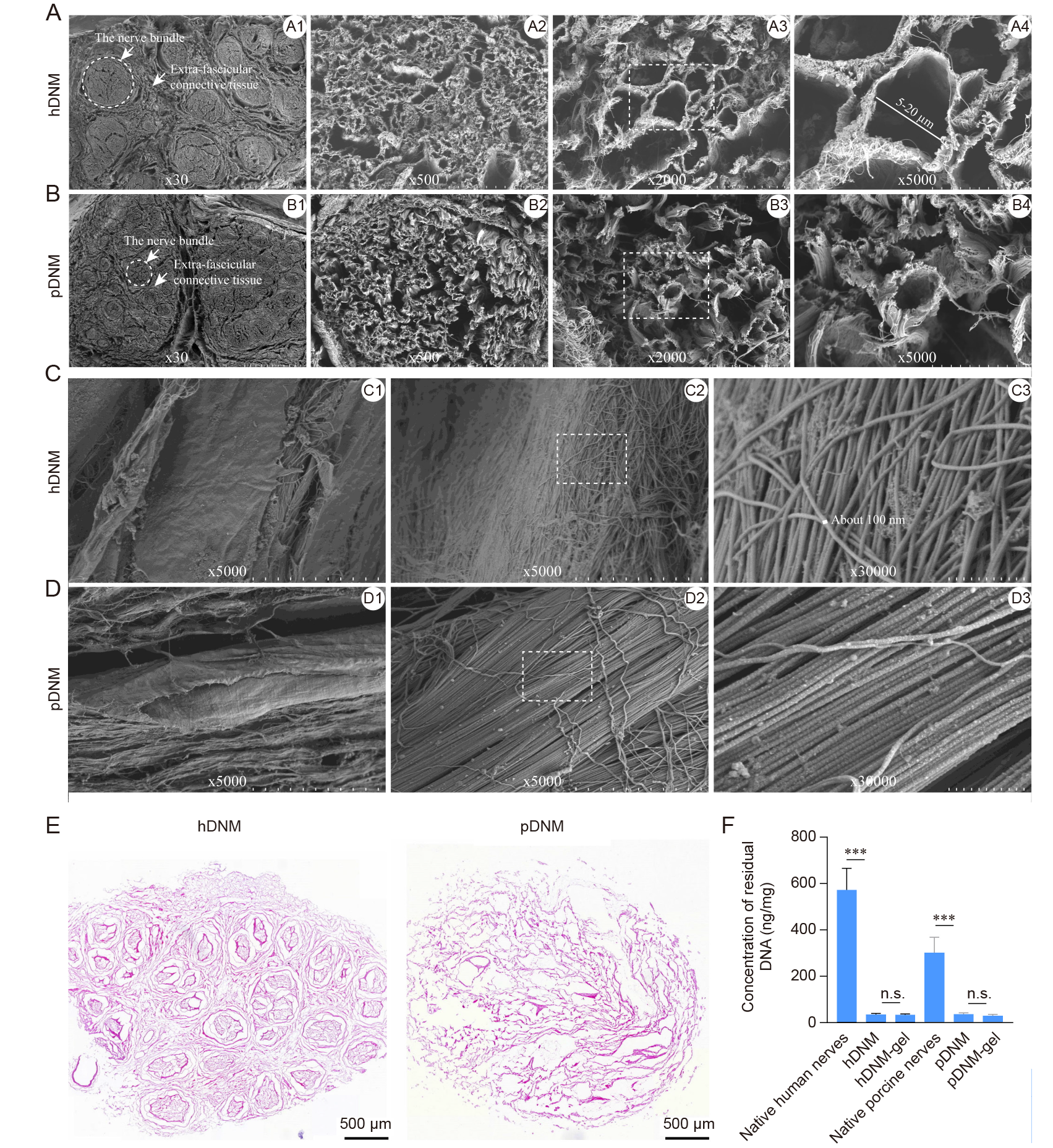
Figure 1. Structural and histological characterisations of the hDNM and pDNM scaffolds. Representative SEM micrographs of the hDNM (A) and pDNM (B) at lower magnification. Representative SEM micrographs of the hDNM (C) and pDNM (D) at higher magnification. Scale bars: 1 mm in A1, B1; 100 μm in A2, B2; 20 μm in A3, B3; 10 μm in A4, B4, C1, C2, D1, D2; and 1 μm in C3, D3. (E) Representative micrographs of hDNM and pDNM cross–sections after H&E staining. Scale bars: 500 μm. (F) DNA content quantified in the fresh tissues, hDNM, pDNM, hDNM–gel and pDNM–gel. Data are expressed as mean ± SD (n = 4). ***P < 0.001. H&E: haematoxylin–eosin; hDNM: human decellularised nerve matrix; n.s: not significant; pDNM: porcine decellularised nerve matrix; SEM: scanning electron microscopy.
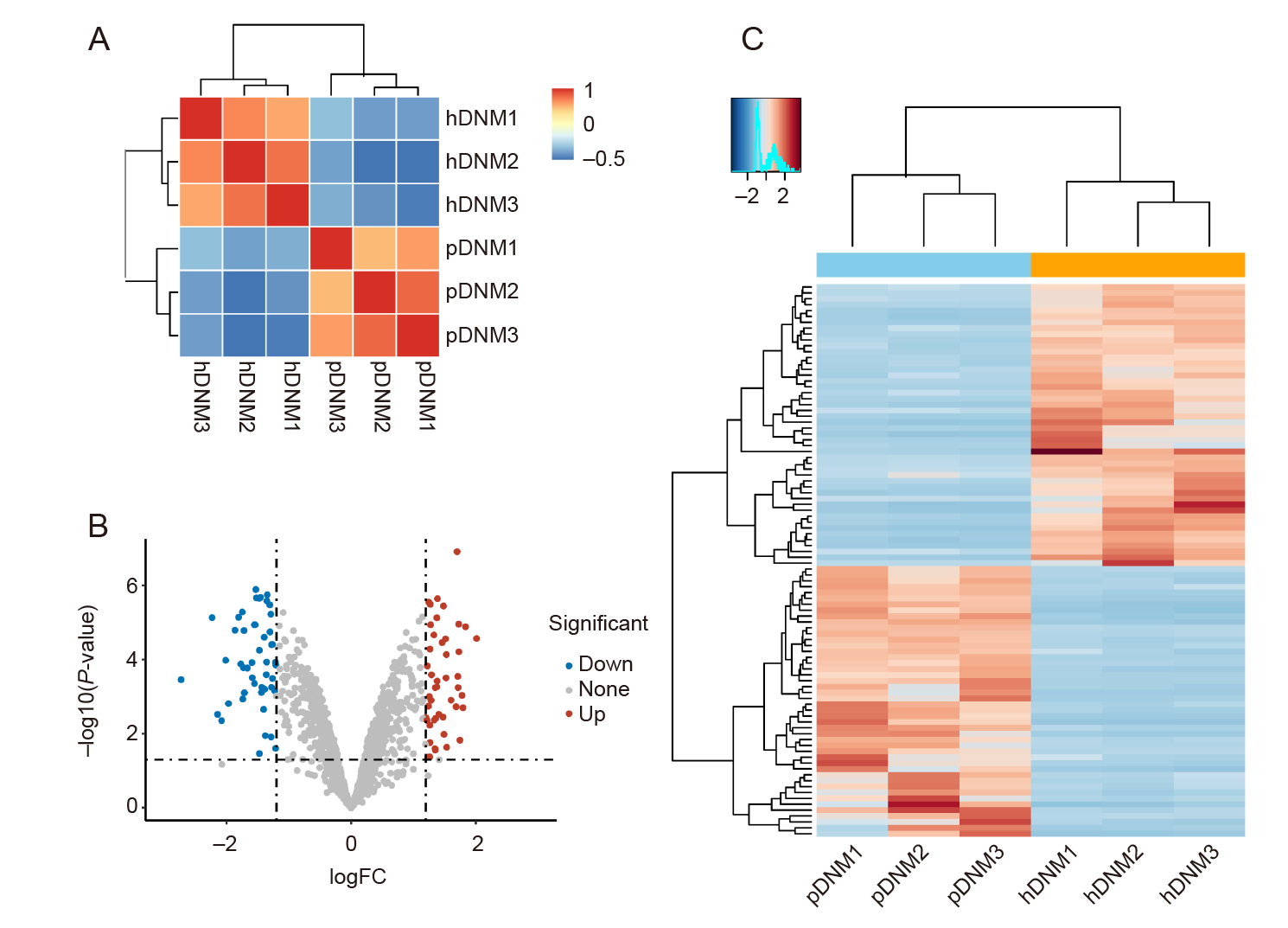
Figure 2. Proteomic analysis of hDNM and pDNM. (A) Unsupervised hierarchical clustering between the hDNM and pDNM using Pearson’s correlation. (B) Volcano plot showing the differentially–expressed proteins between the hDNM and pDNM. (C) Heatmap and cluster dendrogram of protein abundances in the hDNM and pDNM. FC: fold change; hDNM: human decellularised nerve matrix; pDNM: porcine decellularised nerve matrix.
| Classification | pDNM | hDNM | |
|---|---|---|---|
| Core matrisome | ECM glycoproteins | LAMB3, MXRA5, EDIL3, SLIT1, VWA3A, EMILIN3 | EMILIN2, CTGF, LAMA1, SRPX |
| Collagens | None | COL6A6 | |
| Proteoglycans | None | None | |
| ECM–associated proteins | ECM–related proteins | ELFN2, FREM2, C1QL4, PLXNA2, PLXNA4, ANXA9 | GPC5, FCN1, PLXNA3 |
| ECM regulators | ADAMTS14, TIMP3, CTSG, MMP12, MEP1A, ADAMTS7, ADAMTS5 | FAM20C, ADAMTS21, ADAMTS15, ADAMTSL3 | |
| Secreted factors | WNT3A, BMP3, INHBA, MSTN, S100A4, NFSF15, FGF14, S100A13, ANGPTL7, HHIP, BRINP2, GDF3, FGF9 | MEGF11, IL4, GDF5, INHBB |
Table 2. Specific dECM proteins identified only in the pDNM or hDNM
| Classification | pDNM | hDNM | |
|---|---|---|---|
| Core matrisome | ECM glycoproteins | LAMB3, MXRA5, EDIL3, SLIT1, VWA3A, EMILIN3 | EMILIN2, CTGF, LAMA1, SRPX |
| Collagens | None | COL6A6 | |
| Proteoglycans | None | None | |
| ECM–associated proteins | ECM–related proteins | ELFN2, FREM2, C1QL4, PLXNA2, PLXNA4, ANXA9 | GPC5, FCN1, PLXNA3 |
| ECM regulators | ADAMTS14, TIMP3, CTSG, MMP12, MEP1A, ADAMTS7, ADAMTS5 | FAM20C, ADAMTS21, ADAMTS15, ADAMTSL3 | |
| Secreted factors | WNT3A, BMP3, INHBA, MSTN, S100A4, NFSF15, FGF14, S100A13, ANGPTL7, HHIP, BRINP2, GDF3, FGF9 | MEGF11, IL4, GDF5, INHBB |
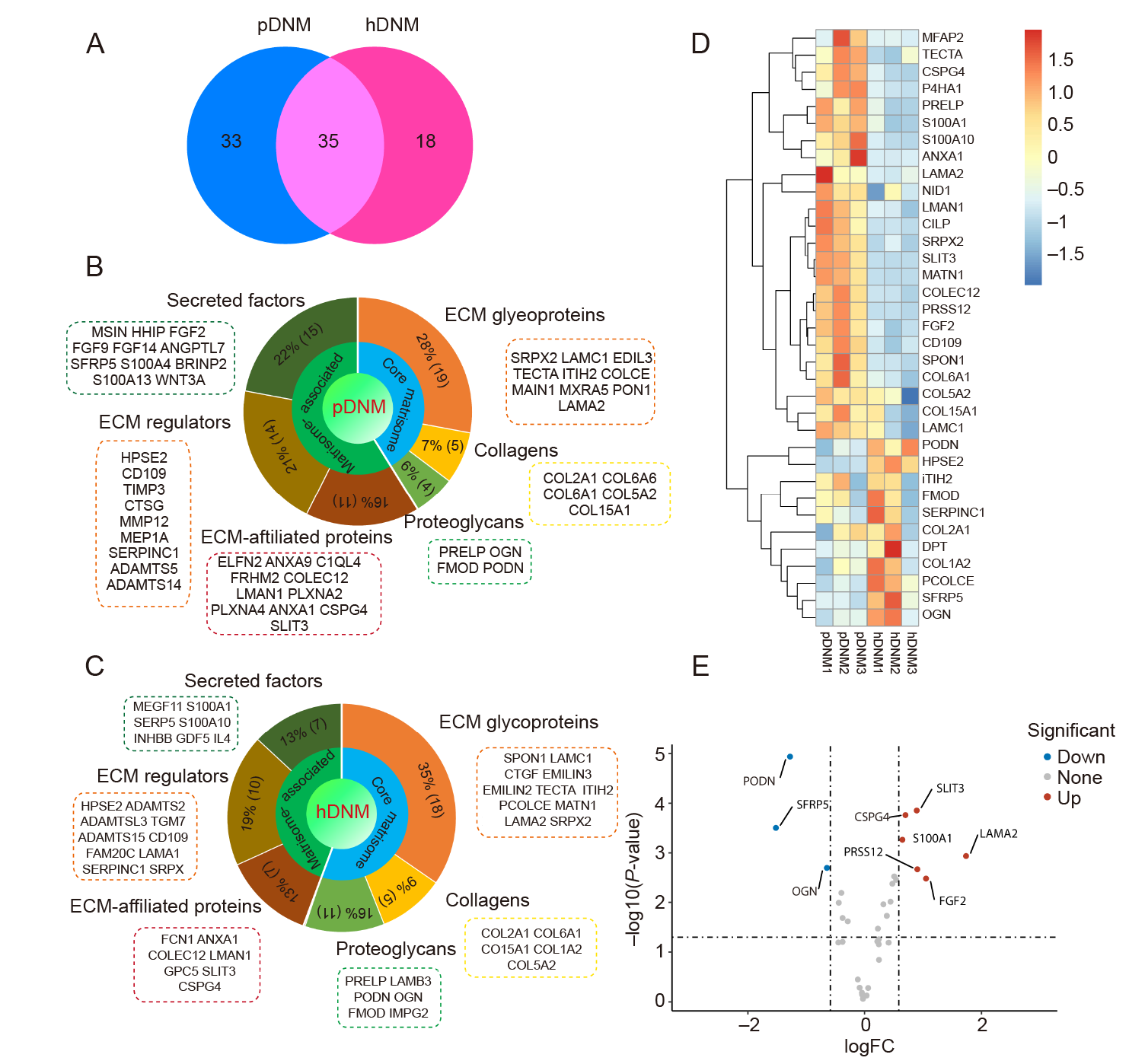
Figure 3. Matrisome analysis of the proteomic composition in pDNM and hDNM. (A) Venn diagram showing the number of ECM proteins detected in hDNM and pDNM. (B, C) Percentages of the ECM proteins and their corresponding matrisome subcategories identified in pDNM (B) and hDNM (C). (D) Heatmap representing significant distinctions in the co–expressed ECM proteins between pDNM and hDNM. The relative abundance of the 35 shared proteins identified in pDNM was higher than that in hDNM. (E) Volcano plot of the differentially expressed ECM proteins in pDNM compared to hDNM. The red and blue dots indicate the significantly up– and down–regulated ECM proteins, respectively (n = 3 for both pDNM and hDNM). ECM: extracellular matrix, hDNM: human decellularised nerve matrix; pDNM: porcine decellularised nerve matrix.
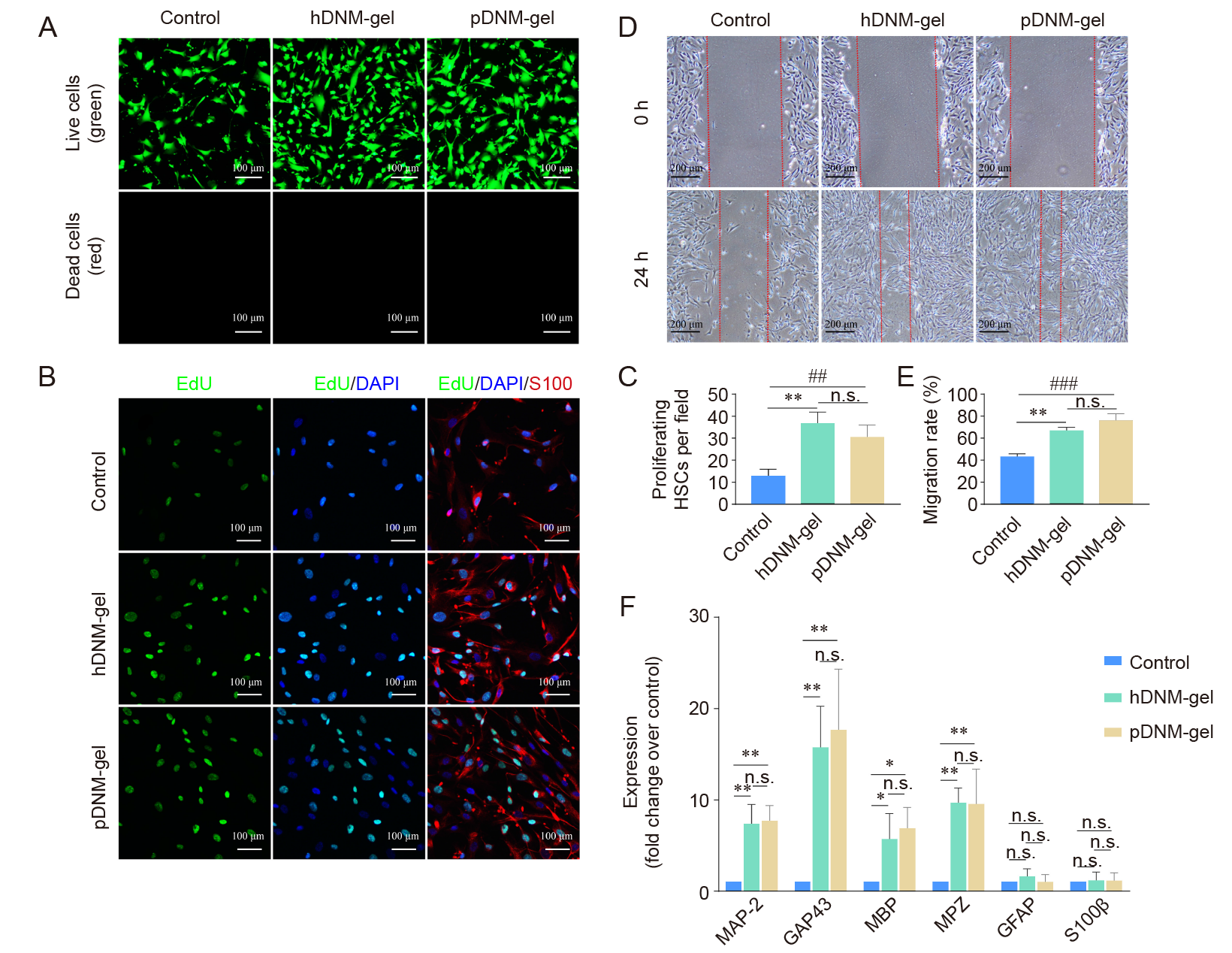
Figure 4. Bioactivities of both hDNM–gel and pDNM–gel in regulating the behaviours of cultured HSCs. (A) Representative fluorescence micrographs of the cultured HSCs on hDNM–gel and pDNM–gel after 48 hours of incubation and live/dead staining, compared to the control (no hydrogel). Scale bars: 100 μm. (B) Representative fluorescence confocal micrographs of HSCs cultured for 48 hours and immunostained with EdU (green), S100 (red), and DAPI (blue). Scale bars: 100 μm. (C) Number of proliferating (EdU+/S100+) HSCs in B. (D) Wound healing characterisation showing the wound gaps at 0 and 24 hours in the control, hDNM–gel, and pDNM–gel groups. Scale bars: 100 μm. (E) HSC migration based on the wound healing experiments in D (n = 3). (F) MAP–2, GAP43, MBP, MPZ, GFAP, and S100β mRNA expression of the HSCs cultured on hDNM–gel and pDNM–gel were significantly upregulated compared to the control group (n = 5). β–actin was used as the reference. Data are shown as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. DAPI: 6–diamidino–2–phenylindole–dihydrochloride; dECM: decellularised extracellular matrix; EdU: 5–ethynyl–2′–deoxyuridine; GAP43: growth–associated protein 43; GFAP: glial fibrillary acidic protein; hDNM–gel: human decellularised nerve matrix hydrogel; HSCs: human Schwann cells; MAP–2: microtubule–associated protein–2; MBP: myelin basic protein; MPZ: myelin protein zero; n.s, not significant; pDNM–gel: porcine decellularised nerve matrix hydrogel.
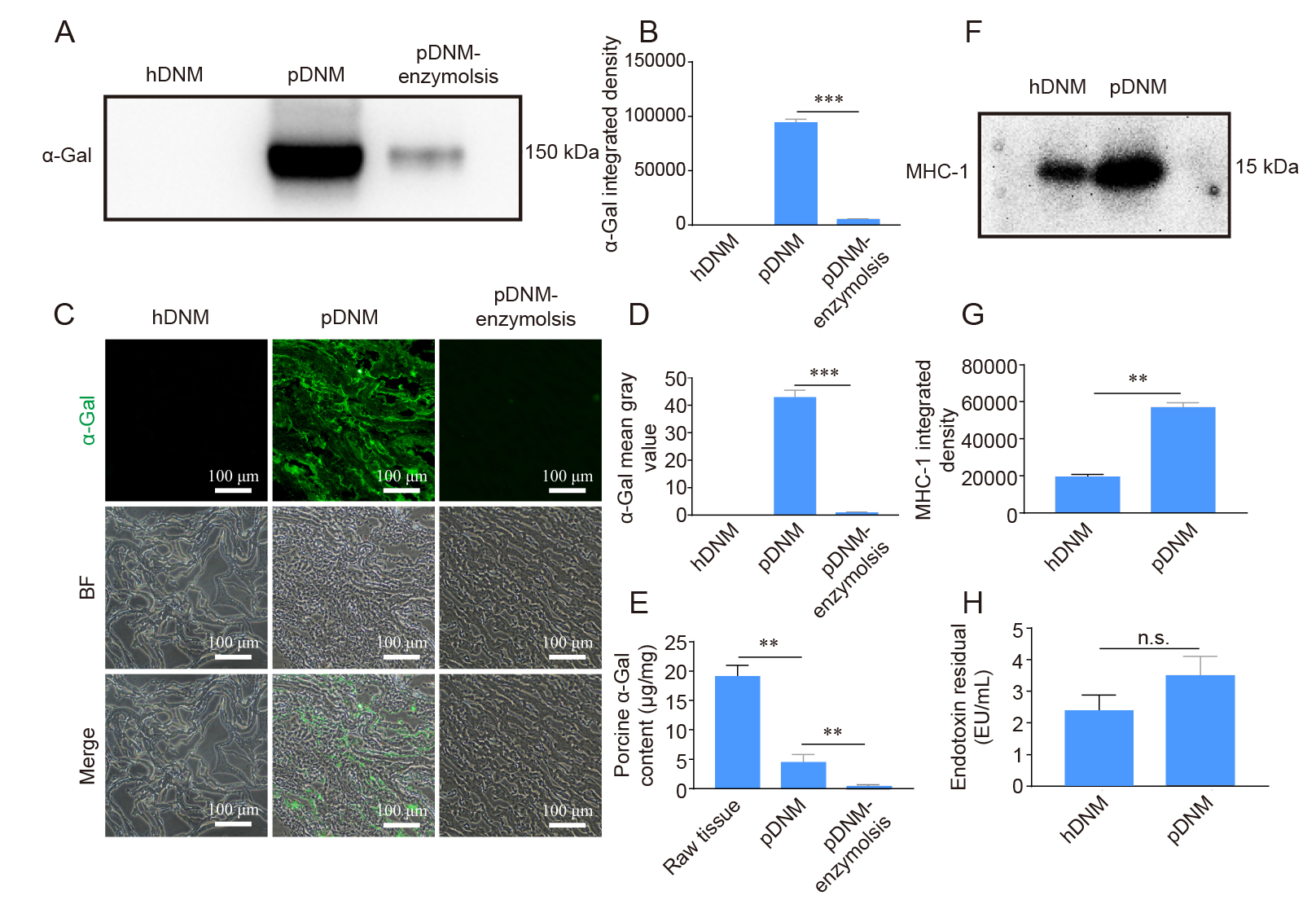
Figure 5. Detection of the immunogenic contents (α–Gal, MHC–1, and endotoxin) in hDNM and pDNM. (A, B) Western blot results and quantification of α–Gal antigen in hDNM, pDNM, and pDNM pre–treated with α–galactosidase (pDNM–enzymolysis). (C) Immunofluorescence staining and BF micrographs showing the presence of α–Gal antigen (green) within the hDNM, pDNM, and pDNM–enzymolysis samples. Scale bars: 100 μm. (D) Quantification of the immunoreactivity of α–Gal antigens. (E) Quantification of α–Gal content in raw porcine nerve tissues, pDNM, and pDNM after α–Gal treatment. (F) Western blot image and (G) quantification of the MHC–1 content in hDNM and pDNM. (H) Quantification of the endotoxin content in hDNM and pDNM. Data are presented as mean ± SD (n = 3). **P < 0.01, ***P < 0.001. α–Gal: α–galactosidase; BF: bright field; hDNM: human decellularised nerve matrix; MHC–1: major histocompatibility complex 1; n.s.: not significant; pDNM: porcine decellularised nerve matrix.
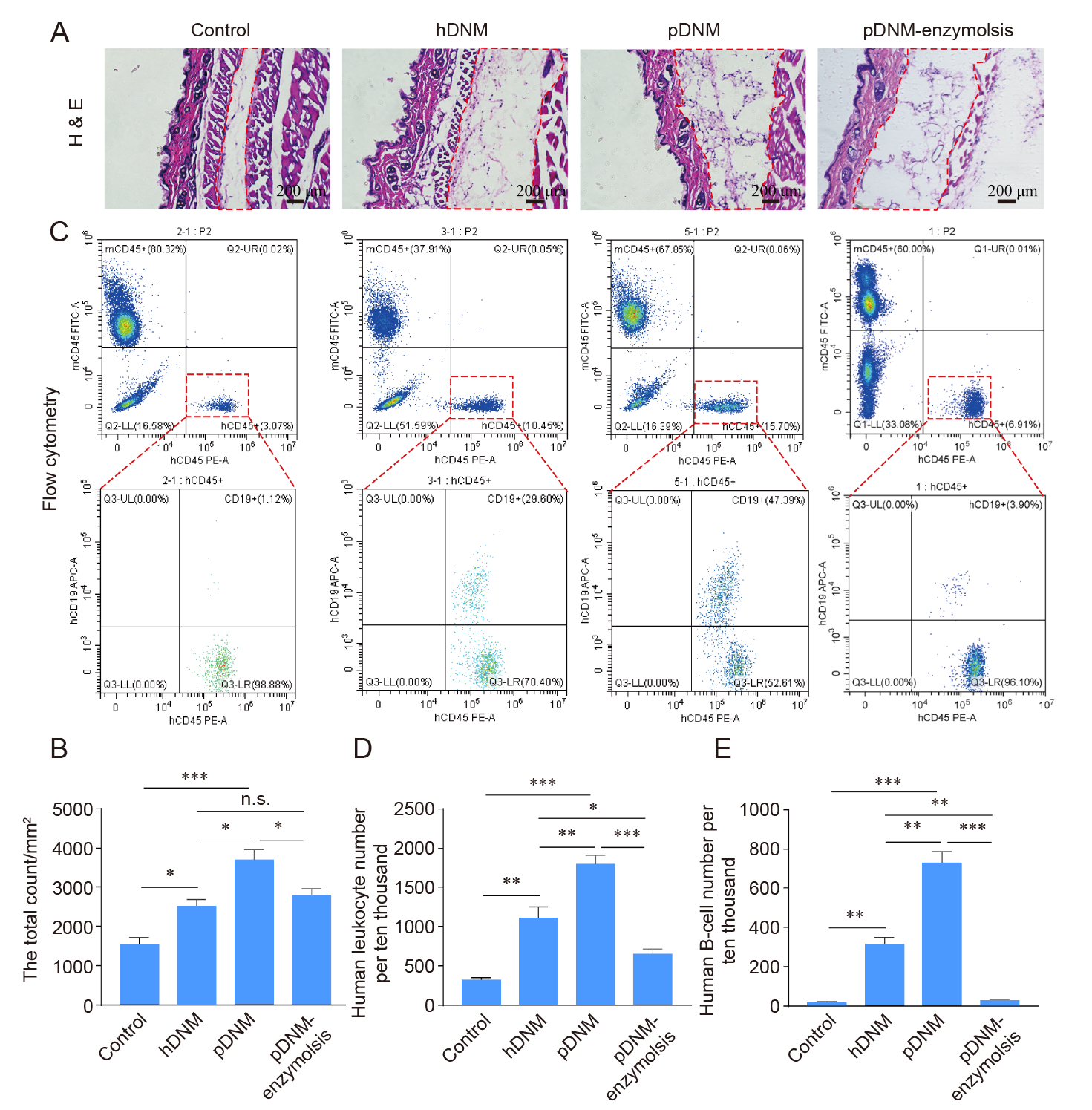
Figure 6. Host immune responses to subcutaneously injected hDNM, pDNM, and pDNM–enzymolysis in a humanised mouse model. Hu–mice received the same volume of sterile saline as the control. (A) H&E staining of the subcutaneous tissues sectioned from the Hu–mice in each group. Scale bars: 200 μm. (B) The total number of infiltrating cells in the dotted box in A based on H&E histological staining. (C) Flow cytometry results showing the human immune cells after treatment. (D, E) Quantitative analysis of the density of human leukocytes (hCD45+) (D) and the density of human B cells (hCD45+ hCD19+) (E) based on flow cytometric assessments. Data are presented as mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. H&E: haematoxylin–eosin; hDNM: human decellularised nerve matrix; n.s: not significant; pDNM: porcine decellularised nerve matrix.
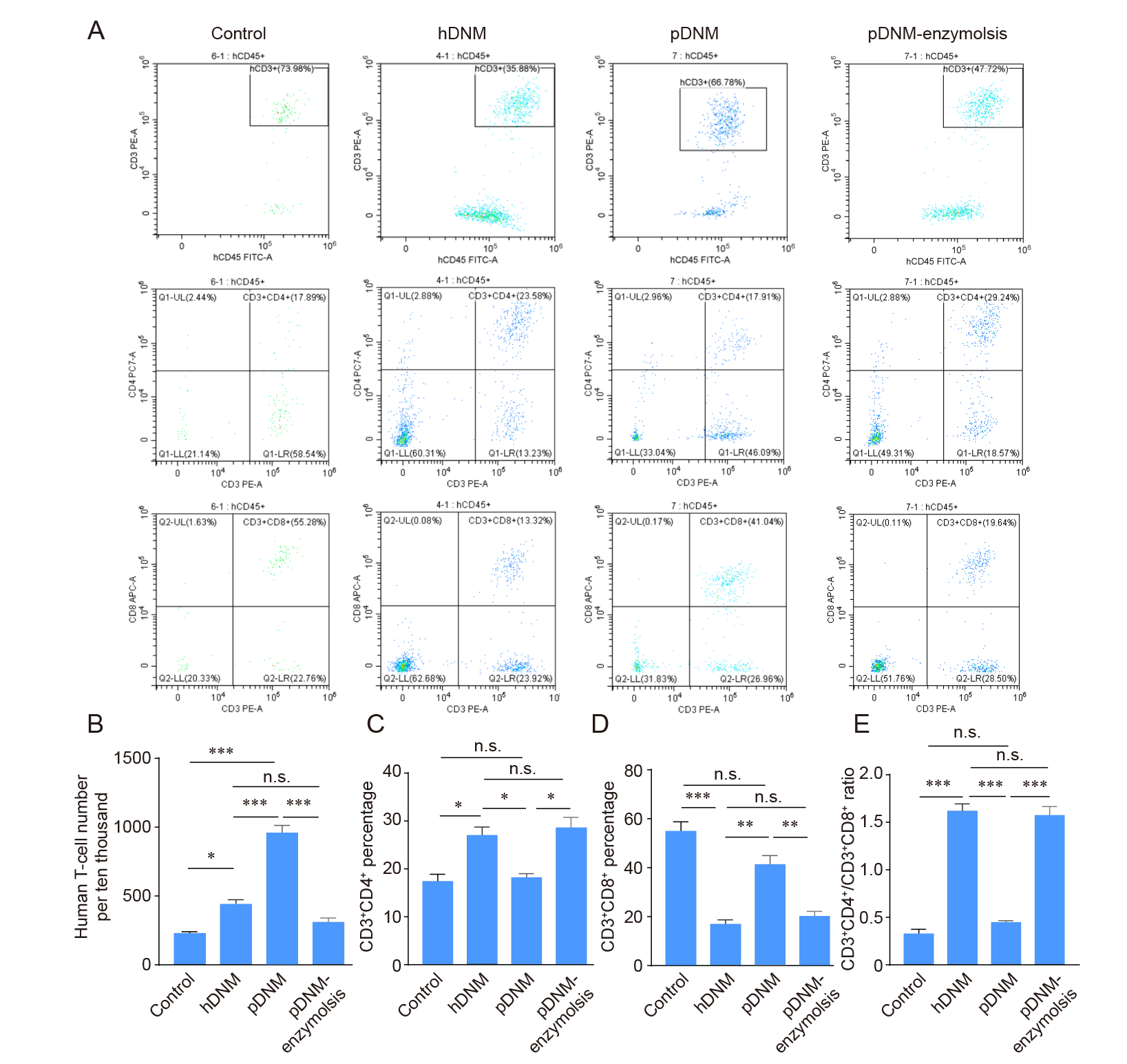
Figure 7. Evaluation of T cell repopulation and their subtypes after injection of hDNM, pDNM, or pDNM–enzymolysis into humanised mice, analysed using flow cytometry. (A) T cells (hCD45+hCD3+) and their subtypes, including T helper cells (hCD3+hCD4+) and cytotoxic T cells (hCD3+hCD8+). (B) The density of total human T cells after introducing the dECMs into Hu–mice. (C) The percentage of the T helper cells (hCD3+hCD4+). (D) The percentage of cytotoxic T cells (hCD3+hCD8+). (E) The ratios of T helper cells/cytotoxic T cells. The data are expressed as mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. hDNM: human decellularised nerve matrix; n.s: not significant; pDNM: porcine decellularised nerve matrix.
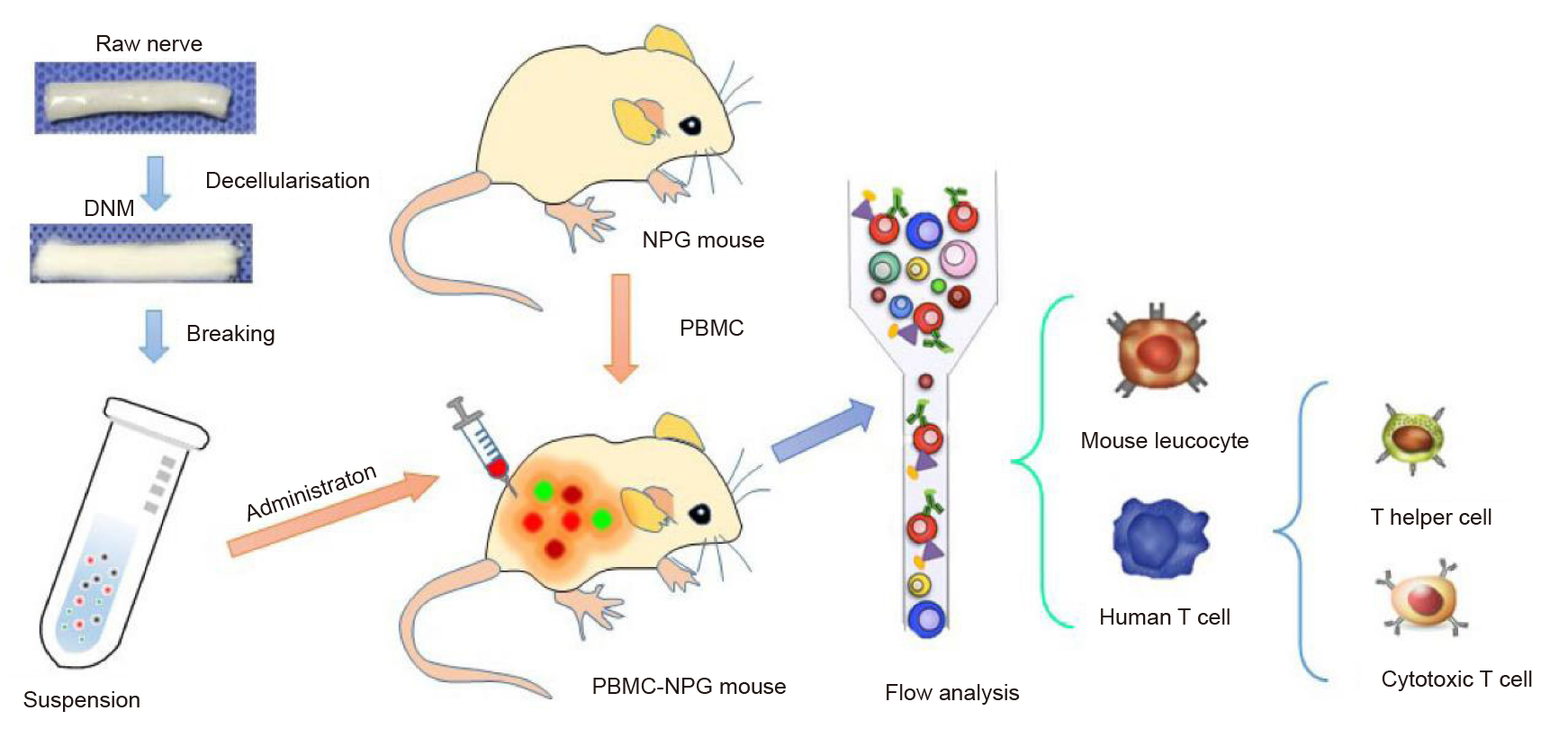
Additional Figure 1. A flow chart of the in vivo study. DNM: decellularised nerve matrix; NPG: NOD–Prkdcscid Il2rgnull; PBMC: peripheral blood mononuclear cell.
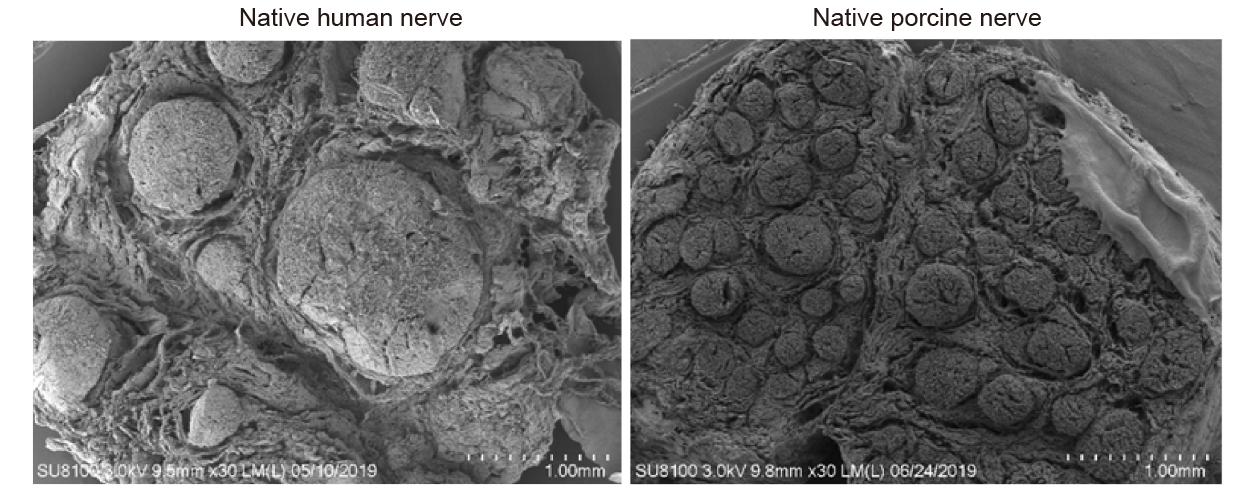
Additional Figure 2. Scanning electron micrographs showing cross–sectional views of a native human nerve and a native porcine nerve. Scale bars: 1 mm.
| 1. | Krishtul, S.; Baruch, L.; Machluf, M. Processed tissue-derived extracellular matrices: tailored platforms empowering diverse therapeutic applications. Adv Funct Mater. 2020, 30, 1900386. |
| 2. |
Garreta, E.; Oria, R.; Tarantino, C.; Pla-Roca, M.; Prado, P.; Fernández-Avilés, F.; Campistol, J. M.; Samitier, J.; Montserrat, N. Tissue engineering by decellularization and 3D bioprinting. Mater Today. 2017, 20, 166-178.
doi: 10.1016/j.mattod.2016.12.005 URL |
| 3. |
Hussey, G. S.; Dziki, J. L.; Badylak, S. F. Extracellular matrix-based materials for regenerative medicine. Nat Rev Mater. 2018, 3, 159-173.
doi: 10.1038/s41578-018-0023-x |
| 4. |
Li, T.; Javed, R.; Ao, Q. Xenogeneic decellularized extracellular matrix-based biomaterials for peripheral nerve repair and regeneration. Curr Neuropharmacol. 2021, 19, 2152-2163.
doi: 10.2174/1570159X18666201111103815 URL |
| 5. |
Zouhair, S.; Sasso, E. D.; Tuladhar, S. R.; Fidalgo, C.; Vedovelli, L.; Filippi, A.; Borile, G.; Bagno, A.; Marchesan, M.; Giorgio, R.; Gregori, D.; Wolkers, W. F.; Romanato, F.; Korossis, S.; Gerosa, G.; Iop, L. A comprehensive comparison of bovine and porcine decellularized pericardia: new insights for surgical applications. Biomolecules. 2020, 10, 371.
doi: 10.3390/biom10030371 URL |
| 6. |
Capella-Monsonís, H.; De Pieri, A.; Peixoto, R.; Korntner, S.; Zeugolis, D. I. Extracellular matrix-based biomaterials as adipose-derived stem cell delivery vehicles in wound healing: a comparative study between a collagen scaffold and two xenografts. Stem Cell Res Ther. 2020, 11, 510.
doi: 10.1186/s13287-020-02021-x |
| 7. |
Bowers, S. L.; Banerjee, I.; Baudino, T. A. The extracellular matrix: at the center of it all. J Mol Cell Cardiol. 2010, 48, 474-482.
doi: 10.1016/j.yjmcc.2009.08.024 URL |
| 8. |
Keane, T. J.; Londono, R.; Turner, N. J.; Badylak, S. F. Consequences of ineffective decellularization of biologic scaffolds on the host response. Biomaterials. 2012, 33, 1771-1781.
doi: 10.1016/j.biomaterials.2011.10.054 URL |
| 9. |
Badylak, S. F. Xenogeneic extracellular matrix as a scaffold for tissue reconstruction. Transpl Immunol. 2004, 12, 367-377.
doi: 10.1016/j.trim.2003.12.016 URL |
| 10. |
Badylak, S. F.; Gilbert, T. W. Immune response to biologic scaffold materials. Semin Immunol. 2008, 20, 109-116.
doi: 10.1016/j.smim.2007.11.003 URL |
| 11. | Kasper, M.; Deister, C.; Beck, F.; Schmidt, C. E. Bench-to-bedside lessons learned: commercialization of an acellular nerve graft. Adv Healthc Mater. 2020, 9, e2000174. |
| 12. | Yang, L. M.; Liu, X. L.; Zhu, Q. T.; Zhang, Y.; Xi, T. F.; Hu, J.; He, C. F.; Jiang, L. Human peripheral nerve-derived scaffold for tissue-engineered nerve grafts: histology and biocompatibility analysis. J Biomed Mater Res B Appl Biomater. 2011, 96, 25-33. |
| 13. | Colwell, A. S.; Longaker, M. T.; Lorenz, H. P. Mammalian fetal organ regeneration. Adv Biochem Eng Biotechnol. 2005, 93, 83-100. |
| 14. |
Sicari, B. M.; Johnson, S. A.; Siu, B. F.; Crapo, P. M.; Daly, K. A.; Jiang, H.; Medberry, C. J.; Tottey, S.; Turner, N. J.; Badylak, S. F. The effect of source animal age upon the in vivo remodeling characteristics of an extracellular matrix scaffold. Biomaterials. 2012, 33, 5524-5533.
doi: 10.1016/j.biomaterials.2012.04.017 URL |
| 15. |
Badylak, S. F. The extracellular matrix as a biologic scaffold material. Biomaterials. 2007, 28, 3587-3593.
doi: 10.1016/j.biomaterials.2007.04.043 URL |
| 16. | Seif-Naraghi, S. B.; Singelyn, J. M.; Salvatore, M. A.; Osborn, K. G.; Wang, J. J.; Sampat, U.; Kwan, O. L.; Strachan, G. M.; Wong, J.; Schup-Magoffin, P. J.; Braden, R. L.; Bartels, K.; DeQuach, J. A.; Preul, M.; Kinsey, A. M.; DeMaria, A. N.; Dib, N.; Christman, K. L. Safety and efficacy of an injectable extracellular matrix hydrogel for treating myocardial infarction. Sci Transl Med. 2013, 5, 173ra125. |
| 17. |
Chen, S.; Du, Z.; Zou, J.; Qiu, S.; Rao, Z.; Liu, S.; Sun, X.; Xu, Y.; Zhu, Q.; Liu, X.; Mao, H. Q.; Bai, Y.; Quan, D. Promoting neurite growth and schwann cell migration by the harnessing decellularized nerve matrix onto nanofibrous guidance. ACS Appl Mater Interfaces. 2019, 11, 17167-17176.
doi: 10.1021/acsami.9b01066 URL |
| 18. |
Zheng, C.; Yang, Z.; Chen, S.; Zhang, F.; Rao, Z.; Zhao, C.; Quan, D.; Bai, Y.; Shen, J. Nanofibrous nerve guidance conduits decorated with decellularized matrix hydrogel facilitate peripheral nerve injury repair. Theranostics. 2021, 11, 2917-2931.
doi: 10.7150/thno.50825 URL |
| 19. |
Deng, R.; Luo, Z.; Rao, Z.; Lin, Z.; Chen, S.; Zhou, J.; Zhu, Q.; Liu, X.; Bai, Y.; Quan, D. Decellularized extracellular matrix containing electrospun fibers for nerve regeneration: a comparison between core-shell structured and preblended composites. Adv Fiber Mater. 2022, 4, 503-519.
doi: 10.1007/s42765-021-00124-5 |
| 20. |
Xu, Y.; Zhou, J.; Liu, C.; Zhang, S.; Gao, F.; Guo, W.; Sun, X.; Zhang, C.; Li, H.; Rao, Z.; Qiu, S.; Zhu, Q.; Liu, X.; Guo, X.; Shao, Z.; Bai, Y.; Zhang, X.; Quan, D. Understanding the role of tissue-specific decellularized spinal cord matrix hydrogel for neural stem/progenitor cell microenvironment reconstruction and spinal cord injury. Biomaterials. 2021, 268, 120596.
doi: 10.1016/j.biomaterials.2020.120596 URL |
| 21. |
Lin, T.; Liu, S.; Chen, S.; Qiu, S.; Rao, Z.; Liu, J.; Zhu, S.; Yan, L.; Mao, H.; Zhu, Q.; Quan, D.; Liu, X. Hydrogel derived from porcine decellularized nerve tissue as a promising biomaterial for repairing peripheral nerve defects. Acta Biomater. 2018, 73, 326-338.
doi: 10.1016/j.actbio.2018.04.001 URL |
| 22. |
Rao, Z.; Lin, T.; Qiu, S.; Zhou, J.; Liu, S.; Chen, S.; Wang, T.; Liu, X.; Zhu, Q.; Bai, Y.; Quan, D. Decellularized nerve matrix hydrogel scaffolds with longitudinally oriented and size-tunable microchannels for peripheral nerve regeneration. Mater Sci Eng C Mater Biol Appl. 2021, 120, 111791.
doi: 10.1016/j.msec.2020.111791 URL |
| 23. |
Keane, T. J.; Badylak, S. F. The host response to allogeneic and xenogeneic biological scaffold materials. J Tissue Eng Regen Med. 2015, 9, 504-511.
doi: 10.1002/term.1874 URL |
| 24. |
Brown, B. N.; Londono, R.; Tottey, S.; Zhang, L.; Kukla, K. A.; Wolf, M. T.; Daly, K. A.; Reing, J. E.; Badylak, S. F. Macrophage phenotype as a predictor of constructive remodeling following the implantation of biologically derived surgical mesh materials. Acta Biomater. 2012, 8, 978-987.
doi: 10.1016/j.actbio.2011.11.031 URL |
| 25. |
Brown, B. N.; Valentin, J. E.; Stewart-Akers, A. M.; McCabe, G. P.; Badylak, S. F. Macrophage phenotype and remodeling outcomes in response to biologic scaffolds with and without a cellular component. Biomaterials. 2009, 30, 1482-1491.
doi: 10.1016/j.biomaterials.2008.11.040 URL |
| 26. |
Galili, U. Interaction of the natural anti-Gal antibody with alpha-galactosyl epitopes: a major obstacle for xenotransplantation in humans. Immunol Today. 1993, 14, 480-482.
doi: 10.1016/0167-5699(93)90261-I URL |
| 27. |
Mestas, J.; Hughes, C. C. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004, 172, 2731-2738.
doi: 10.4049/jimmunol.172.5.2731 URL |
| 28. |
Qiu, S.; Rao, Z.; He, F.; Wang, T.; Xu, Y.; Du, Z.; Yao, Z.; Lin, T.; Yan, L.; Quan, D.; Zhu, Q.; Liu, X. Decellularized nerve matrix hydrogel and glial-derived neurotrophic factor modifications assisted nerve repair with decellularized nerve matrix scaffolds. J Tissue Eng Regen Med. 2020, 14, 931-943.
doi: 10.1002/term.v14.7 URL |
| 29. | Devaud, Y. R.; Avilla-Royo, E.; Trachsel, C.; Grossmann, J.; Martin, I.; Lutolf, M. P.; Ehrbar, M. Label-free quantification proteomics for the identification of mesenchymal stromal cell matrisome inside 3D poly(ethylene glycol) hydrogels. Adv Healthc Mater. 2018, 7, e1800534. |
| 30. |
UniProt, Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480-D489.
doi: 10.1093/nar/gkaa1100 URL |
| 31. |
Morfeld, P. Controlling the false discovery rate in many SMR analyses. J Occup Environ Med. 2016, 58, e21-22.
doi: 10.1097/JOM.0000000000000599 URL |
| 32. |
Naba, A.; Clauser, K. R.; Ding, H.; Whittaker, C. A.; Carr, S. A.; Hynes, R. O. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 2016, 49, 10-24.
doi: 10.1016/j.matbio.2015.06.003 URL |
| 33. | Schneider, C. A.; Rasband, W. S.; Eliceiri, K. W. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012, 9, 671-675. |
| 34. |
Bubner, B.; Baldwin, I. T. Use of real-time PCR for determining copy number and zygosity in transgenic plants. Plant Cell Rep. 2004, 23, 263-271.
doi: 10.1007/s00299-004-0859-y URL |
| 35. |
Stone, K. R.; Ayala, G.; Goldstein, J.; Hurst, R.; Walgenbach, A.; Galili, U. Porcine cartilage transplants in the cynomolgus monkey. III. Transplantation of alpha-galactosidase-treated porcine cartilage. Transplantation. 1998, 65, 1577-1583.
doi: 10.1097/00007890-199806270-00007 URL |
| 36. |
Percie du Sert, N.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M. T.; Baker, M.; Browne, W. J.; Clark, A.; Cuthill, I. C.; Dirnagl, U.; Emerson, M.; Garner, P.; Holgate, S. T.; Howells, D. W.; Karp, N. A.; Lazic, S. E.; Lidster, K.; MacCallum, C. J.; Macleod, M.; Pearl, E. J.; Petersen, O. H.; Rawle, F.; Reynolds, P.; Rooney, K.; Sena, E. S.; Silberberg, S. D.; Steckler, T.; Würbel, H. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biol. 2020, 18, e3000410.
doi: 10.1371/journal.pbio.3000410 URL |
| 37. | Lan, P.; Tonomura, N.; Shimizu, A.; Wang, S.; Yang, Y. G. Reconstitution of a functional human immune system in immunodeficient mice through combined human fetal thymus/liver and CD34+ cell transplantation. Blood. 2006, 108, 487-492. |
| 38. | Khan, S.; Kaihara, K. A. Single-cell RNA-sequencing of peripheral blood mononuclear cells with ddSEQ. Methods Mol Biol. 2019, 1979, 155-176. |
| 39. |
Wang, R. M.; Johnson, T. D.; He, J.; Rong, Z.; Wong, M.; Nigam, V.; Behfar, A.; Xu, Y.; Christman, K. L. Humanized mouse model for assessing the human immune response to xenogeneic and allogeneic decellularized biomaterials. Biomaterials. 2017, 129, 98-110.
doi: 10.1016/j.biomaterials.2017.03.016 URL |
| 40. |
Crapo, P. M.; Medberry, C. J.; Reing, J. E.; Tottey, S.; van der Merwe, Y.; Jones, K. E.; Badylak, S. F. Biologic scaffolds composed of central nervous system extracellular matrix. Biomaterials. 2012, 33, 3539-3547.
doi: 10.1016/j.biomaterials.2012.01.044 URL |
| 41. | Behan, B. L.; DeWitt, D. G.; Bogdanowicz, D. R.; Koppes, A. N.; Bale, S. S.; Thompson, D. M. Single-walled carbon nanotubes alter Schwann cell behavior differentially within 2D and 3D environments. J Biomed Mater Res A. 2011, 96, 46-57. |
| 42. |
Aamodt, J. M.; Grainger, D. W. Extracellular matrix-based biomaterial scaffolds and the host response. Biomaterials. 2016, 86, 68-82.
doi: 10.1016/j.biomaterials.2016.02.003 URL |
| 43. |
Walsh, N. C.; Kenney, L. L.; Jangalwe, S.; Aryee, K. E.; Greiner, D. L.; Brehm, M. A.; Shultz, L. D. Humanized mouse models of clinical disease. Annu Rev Pathol. 2017, 12, 187-215.
doi: 10.1146/pathmechdis.2017.12.issue-1 URL |
| 44. |
Kočí, Z.; Výborný, K.; Dubišová, J.; Vacková, I.; Jäger, A.; Lunov, O.; Jiráková, K.; Kubinová, Š. Extracellular matrix hydrogel derived from human umbilical cord as a scaffold for neural tissue repair and its comparison with extracellular matrix from porcine tissues. Tissue Eng Part C Methods. 2017, 23, 333-345.
doi: 10.1089/ten.tec.2017.0089 URL |
| 45. |
Tan, Q. W.; Zhang, Y.; Luo, J. C.; Zhang, D.; Xiong, B. J.; Yang, J. Q.; Xie, H. Q.; Lv, Q. Hydrogel derived from decellularized porcine adipose tissue as a promising biomaterial for soft tissue augmentation. J Biomed Mater Res A. 2017, 105, 1756-1764.
doi: 10.1002/jbm.a.v105.6 URL |
| 46. |
Bikhet, M.; Morsi, M.; Hara, H.; Rhodes, L. A.; Carlo, W. F.; Cleveland, D.; Cooper, D. K. C.; Iwase, H. The immune system in infants: Relevance to xenotransplantation. Pediatr Transplant. 2020, 24, e13795.
doi: 10.1111/petr.v24.7 URL |
| 47. |
Yan, L.; Guo, Y.; Qi, J.; Zhu, Q.; Gu, L.; Zheng, C.; Lin, T.; Lu, Y.; Zeng, Z.; Yu, S.; Zhu, S.; Zhou, X.; Zhang, X.; Du, Y.; Yao, Z.; Lu, Y.; Liu, X. Iodine and freeze-drying enhanced high-resolution MicroCT imaging for reconstructing 3D intraneural topography of human peripheral nerve fascicles. J Neurosci Methods. 2017, 287, 58-67.
doi: 10.1016/j.jneumeth.2017.06.009 URL |
| 48. |
Spang, M. T.; Christman, K. L. Extracellular matrix hydrogel therapies: in vivo applications and development. Acta Biomater. 2018, 68, 1-14.
doi: 10.1016/j.actbio.2017.12.019 URL |
| 49. |
Bi, H.; Ye, K.; Jin, S. Proteomic analysis of decellularized pancreatic matrix identifies collagen V as a critical regulator for islet organogenesis from human pluripotent stem cells. Biomaterials. 2020, 233, 119673.
doi: 10.1016/j.biomaterials.2019.119673 URL |
| 50. |
Choudhury, D.; Yee, M.; Sheng, Z. L. J.; Amirul, A.; Naing, M. W. Decellularization systems and devices: State-of-the-art. Acta Biomater. 2020, 115, 51-59.
doi: 10.1016/j.actbio.2020.07.060 URL |
| 51. |
Xing, H.; Lee, H.; Luo, L.; Kyriakides, T. R. Extracellular matrix-derived biomaterials in engineering cell function. Biotechnol Adv. 2020, 42, 107421.
doi: 10.1016/j.biotechadv.2019.107421 URL |
| 52. |
Yue, Y.; Xu, W.; Kan, Y.; Zhao, H. Y.; Zhou, Y.; Song, X.; Wu, J.; Xiong, J.; Goswami, D.; Yang, M.; Lamriben, L.; Xu, M.; Zhang, Q.; Luo, Y.; Guo, J.; Mao, S.; Jiao, D.; Nguyen, T. D.; Li, Z.; Layer, J. V.; Li, M.; Paragas, V.; Youd, M. E.; Sun, Z.; Ding, Y.; Wang, W.; Dou, H.; Song, L.; Wang, X.; Le, L.; Fang, X.; George, H.; Anand, R.; Wang, S. Y.; Westlin, W. F.; Güell, M.; Markmann, J.; Qin, W.; Gao, Y.; Wei, H. J.; Church, G. M.; Yang, L. Extensive germline genome engineering in pigs. Nat Biomed Eng. 2021, 5, 134-143.
doi: 10.1038/s41551-020-00613-9 |
| 53. |
Niu, D.; Wei, H. J.; Lin, L.; George, H.; Wang, T.; Lee, I. H.; Zhao, H. Y.; Wang, Y.; Kan, Y.; Shrock, E.; Lesha, E.; Wang, G.; Luo, Y.; Qing, Y.; Jiao, D.; Zhao, H.; Zhou, X.; Wang, S.; Wei, H.; Güell, M.; Church, G. M.; Yang, L. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science. 2017, 357, 1303-1307.
doi: 10.1126/science.aan4187 URL |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||