Introduction
Coronaviruses belong to the family Coronaviridae, subfamily Coronavirinae that can infect a wide variety of animals, including humans, bats, camels, and birds, and cause respiratory diseases. In December 2019, clusters of patients with pneumonia-like symptoms were admitted to the hospitals, which were later reported to be caused by a novel coronavirus (severe acute respiratory syndrome coronavirus 2, SARS-CoV-2). By January 30, 2020, the outbreak had been declared a public health emergency of international concern by the World Health Organization.1 The global pandemic coronavirus disease 2019 (COVID-19) outbreak had caused > 47 million confirmed infections and > 1.2 million deaths by November 2020.2 Additionally, the virus was reported to be transmitted from human to human by close contact. Currently, rapid and accurate diagnosis is essential for the prevention of extensive COVID-19 outbreaks.
Implementation of effective screening procedures and diagnosis of infected individuals is indispensable for disease management during epidemics. Current diagnostic tests for SARS-CoV-2 include real-time reverse transcriptase-polymerase chain reaction-based assays which require high-end facilities and considerable time.3, 4 In light of the rapid spread of the disease, the current diagnosis based on molecular tests is not field-usable, is expensive, and is not rapid enough to screen infected individuals. In addition the results might vary based on the viral load and quality of the sample.5 There is great interest in using point-of-care serology tests that are easily portable, easy to use, and cost-effective, to overcome these limitations. Lateral flow immunoassays (LFIAs), or dipstick tests, offer all these advantages. Like in-home pregnancy tests, LFIAs are single-use point-of-care diagnostic tests that can be used outside a laboratory.6 In COVID-19, specific IgM antibodies are produced early in the infection, whereas specific IgG antibodies develop in the later stages.7 Hence, the detection of IgM/IgG antibodies against SARS-CoV-2 using LFIA could help assess the epidemiology of the disease and evaluate the infection and immune status of the human population.8
The receptor-binding domain (RBD) of the spike protein located on the surface of SARS-CoV-2 is responsible for viral entry into the host cell and eliciting the neutralizing antibody response; hence it is considered a promising antigen to use as a vaccine candidate and also for the serological diagnosis of COVID-19.9, 10 Production of recombinant RBD protein to use as a diagnostic reagent in LFIA could be expensive in mammalian or insect cell cultures, representing the major drawback for large-scale diagnostic tests, especially for low-income countries. A practical, accurate diagnostic test can be affordable only if the reagents used for testing are cost-effective. Likewise, if the expression platform produces recombinant RBD protein rapidly at a low cost, the cost of an LFIA would be reduced significantly.
Recently, plants have attracted attention as a potential alternative means of producing recombinant proteins over other traditional expression systems due to the system’s affordability, reliability, scalability, and flexibility. Recent advances in the development of transient expression systems in plants offer rapid, large-scale methods of producing biopharmaceuticals that can be used either as diagnostic or therapeutic agents.11-18 Considering the advantages of the plant expression platform, we expressed recombinant RBD antigen in Nicotiana benthamiana.19 The purified plant-produced antigen was then used to develop a diagnostic reagent to establish and validate the LFIA.
Considering the speed of the spread of the virus and the urgent need for a rapid, cost-effective method to screen large populations, we developed and evaluated a rapid LFIA using plant-produced recombinant RBD protein to detect IgM and IgG antibodies against SARS-CoV-2 in clinical samples from patients in Thailand. We tested and validated the efficacy of the assay using serum from patients in whom real-time reverse transcriptase-polymerase chain reaction confirmed SARS-CoV-2 infection.
Methods
Expression of SARS-CoV-2 RBD in N. benthamiana
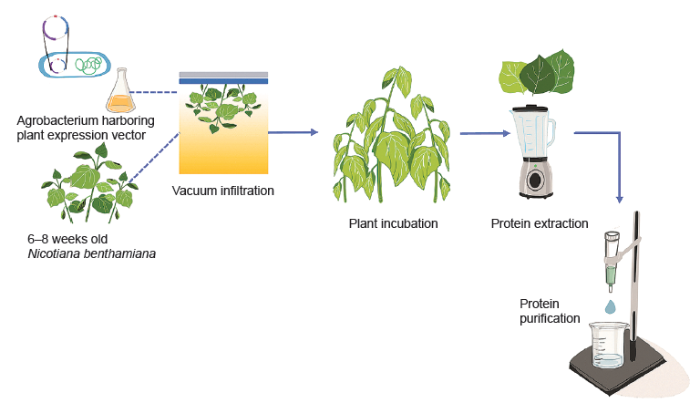
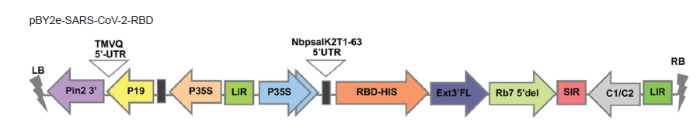
The RBD of SARS-CoV-2 spike protein was transiently expressed in N. benthamiana plants and purified by affinity column chromatography. The procedures were performed as previously reported.19 The schematic representation of recombinant protein expression in plants was shown in Figure 1. Briefly, the gene encoding the SARS-CoV-2 RBD construct was incorporated into a geminiviral vector (pBY2e) (Figure 2). The pBY2e was obtained from Professor Hugh Mason (Arizona State University, USA). The recombinant pBY2e-SARS-CoV-2-RBD vector was transformed into Agrobacterium tumefaciens strain GV3101 (Gold Biotechnology® Inc., Olivette, MO, USA), which was then used to infiltrate N. benthamiana. The SARS-CoV-2 RBD was harvested and purified from N. benthamiana using Ni-nitriloacetic acid affinity resin (Expedeon, Cambridge, UK).
Figure 1.
Figure 1.
Schematic representation for the transient expression of SARS-CoV-2 RBD protein in plants. RBD: receptor-binding domain; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
Figure 2.
Figure 2.
Schematic diagram of the T-DNA region of the geminiviral vector used in this study. pBY2e: geminiviral vector; RBD: receptor-binding domain; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; UTR: untranslated region.
Conjugation of colloidal gold nanoparticles with detecting reagents
The RBD and chicken IgY were conjugated with colloidal gold nanoparticles (AuNPs) and deposited at the conjugate pad as the detecting reagent. AuNPs, 40 nm in diameter, were obtained from Kestrel BioSciences Thailand Co. Ltd. (Pathumthani Province, Thailand). The pH of the AuNP suspension was adjusted to pH 8.0 with 0.2 M K2CO3. The plant-produced SARS-CoV-2 recombinant protein and chicken IgY (10 µg each, Kestrel BioSciences Thailand Co. Ltd.) were then both added to 1 mL AuNP colloid. After incubation for 10 minutes at room temperature, 10% (w/v) bovine serum albumin (0.1 mL, HiMedia Laboratories, Mumbai, India) was added to the mixture to block the AuNP surface. After incubation for 3 hours at room temperature, the mixture was kept at 4°C overnight. Next day, the AuNP-RBD-IgY conjugates were recovered after centrifuge, 9660 × g, 30 minutes, 4°C. The supernatant was discarded, and 1 mL of 1 mg/mL bovine serum albumin in borate buffer was added to the AuNP conjugate and re-suspended. Before dispensing to the conjugate pad, the AuNP-RBD-IgY conjugates were supplemented with 10% (w/v) sucrose and 5% (w/v) trehalose. The AuNP-RBD-IgY conjugates were then sprayed onto the conjugate pad at a rate of 10 μL/cm, then the conjugate pad was dried at 37°C for one hour. The resultant conjugate pad was stored at room temperature, in a dry place, and protected from light.
Preparation and assembly of the LFIA
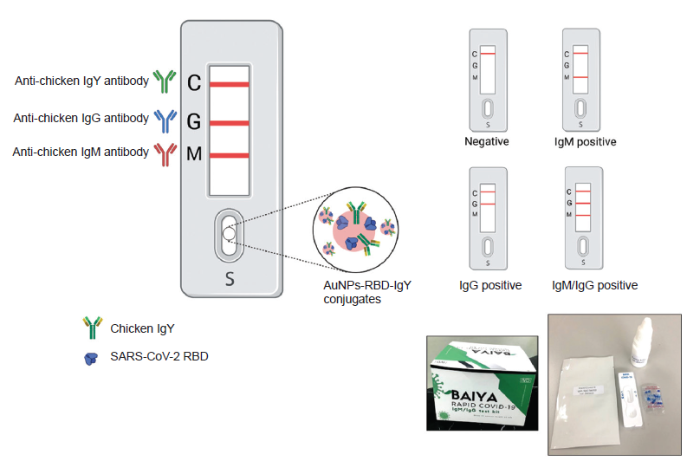
The LFIA framework was demonstrated (Figure 3). The goat anti-human-IgM (1 mg/mL; Kestrel BioSciences Thailand Co. Ltd.), goat anti-human-IgG (1 mg/mL; Lampire Biological Laboratories, Pipersville, PA, USA), and goat anti-chicken IgY (1 mg/mL, Lampire Biological Laboratories) antibodies were immobilized on the IgM test line (M), IgG test line (G), and control line (C), as appropriate. The CN 140 backed nitrocellulose membrane (Unisart®, Sartorius Stedim Biotech GmbH, Goettingen, Germany) was striped with the abovementioned antibodies, at a rate of 0.1 μL/cm. The striped membrane was dried in a convection oven (Memmert UN160 Universal Laboratory, East Troy, WI, USA) at 37°C for 1 hour. The striped membrane was then assembled together with an AuNP-RBD-IgY conjugate pad, sample pad (Ahlstrom 1662, Ahlstrom-Munksjö CytoSep® 1662, Helsinki, Finland), and wick pad (Whatman grade 470, Sigma-Aldrich, St. Louis, MO, USA). Finally, the LFIA was assembled and defined as the Baiya ‘Rapid COVID-19’ IgM/IgG test kit.
Figure 3.
Figure 3.
Schematic illustration of the developed Baiya’ Rapid COVID-19’ IgM/IgG test kit. Different test results are shown. AuNPs: gold nanoparticles; C: control line; G: test line for human IgG; M: test line for human IgM; RBD: receptor-binding domain; S: Sample pad; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
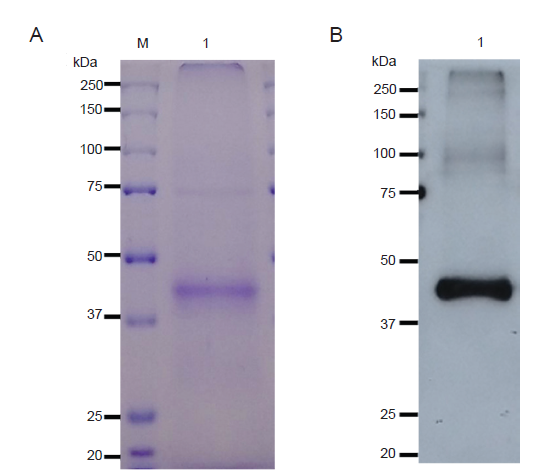
The purified SARS-CoV-2 RBD was tested by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (Figure 4). The bands of protein were detected visually using InstantBlue® (Expedeon, Cambridge, UK) staining. The specific analysis of SARS-CoV-2 RBD was done via western blotting. After transferring proteins from sodium dodecyl sulfate-polyacrylamide gel electrophoresis to a nitrocellulose membrane (Bio-Rad Laboratories, Inc., Hercules, CA, USA), the membrane was treated with skim milk solution. The SARS-CoV-2 RBD was probed with anti-His antibody (HRP conjugate, ab1187, Abcam, Cambridge, UK). Finally, SARS-CoV-2 RBD was detected with enhanced chemiluminescence plus detection reagent (Abcam).
Figure 4.
Figure 4.
Western blot analysis of purified SARS-CoV-2 RBD protein from Nicotiana benthamiana agroinfiltrated with pBY2e-SARS-CoV-2-RBD. (A, B) The purified SARS-CoV-2 RBD protein was loaded at 4 µg/lane under reducing conditions and visualized with InstantBlue® (A) The purified SARS-CoV-2 RBD protein was loaded at 200 ng/lane under reducing conditions and detected with a horseradish peroxidase-conjugated rabbit anti-His antibody (B). M represents the protein molecular weight ladder, and lane 1 shows the purified SARS-CoV-2 RBD. pBY2e: geminiviral vector; RBD: receptor-binding domain; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
Testing of COVID-19 samples using the LFIA system
Serum samples from SARS-CoV-2-infected patients (20 males and 31 females with averages ages of 33 and 42 years, respectively) and negative serum samples were tested using the developed Baiya’ Rapid COVID-19’ IgM/IgG test kit. The Ethics Committee (the Faculty of Medicine, Chulalongkorn University) approved this human sample research (COA No. 354/2020 and IRB No. 236/63) since March 20, 2020. The LFIA strip was taken from the sealed pouch immediately before use. Ten microliters of a patient’s serum were added onto the sample-loading area, followed by two drops of phosphate-buffered saline with 0.05% (v/v) Tween-20. The results were read visually and interpreted 15 minutes after testing. The 51 serum samples from patients confirmed with SARS-CoV-2 infection were tested to evaluate the sensitivity of the LFIA system. The specificity of the LFIA was evaluated using 150 residual serum samples presumed negative for SARS-CoV-2 infection, which were obtained from the Thai Red Cross blood bank. The specificity and sensitivity of the rapid test kits were calculated as follows:
Sensitivity (%) = True positive/(True positive + False negative) × 100
Specificity (%) = True negative/(True negative + False positive)× 100
Results
Expression and purification of the SARS-CoV-2 RBD
Expression of the SARS-CoV-2 RBD in N. benthamiana was successfully achieved. As expected from the corresponding amino acid sequence, the purified SARS-CoV-2 RBD was observed to have a molecular weight of approximately 38 kDa. The SARS-CoV-2 RBD was applied as a diagnostic reagent for the COVID-19 LFIA .
Testing of COVID-19 samples using the LFIA system
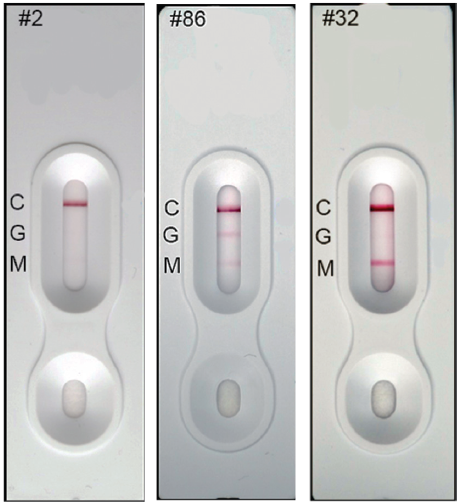
The Baiya ‘Rapid COVID-19’ IgM/IgG test kit was designed and manufactured by Baiya Phytopharm Co., Ltd., Bangkok, Thailand. The test kit was made available in cassette form. The protocol was based on a lateral flow qualitative immunoassay for the rapid and simultaneous detection of anti-SARS-CoV-2 IgM and anti-SARS-CoV-2-IgG antibodies in human samples. When a patient is infected with SARS-CoV-2, anti-SARS-CoV-2 IgM and anti-SARS-CoV-2-IgG are produced. The antibodies used reacted with the RBD of AuNP-RBD-IgY conjugates in the conjugate pad. Then the AuNP-RBD-IgY conjugates bound to anti-SARS-CoV-2 IgM/IgG antibodies were captured by the antibody deposited on the test line, resulting in the appearance of a red line. If a serum sample was negative for anti-SARS-CoV-2 IgM/IgG antibodies, the test line(s) would not be observed. The control line was always observed because AuNP-RBD-IgY conjugates could be captured by the anti-IgY antibody deposited as the control line. The Baiya ‘Rapid COVID-19’ rapid IgM/IgG test kit is shown in Figure 3. A total of three detection lines are present on the strip, the IgM (M), IgG (G), and control (C) lines. Development of the red line in the control position shows that the test is valid. If no colour develops on the control line, the test is invalid, and the test should be repeated with another cassette. The schematic interpretation of the result is presented in Figure 3. Representative test strips showing the results of serum samples collected from different Thai patients are shown in Figure 5. The developed IgM/IgG test kit has a sensitivity and specificity of 94.1% (48/51) and 98% (148/150), respectively. The performance of the LFIA using plant-made SARS-CoV-2 RBD is similar to those achieved with SARS-CoV-2 RBD produced using other expression systems.20, 21 Overall, the plant-based SARS-CoV-2 RBD was effective for the development of a COVID-19 diagnostic assay.
Figure 5.
Figure 5.
Representative test strips showing the results of serum samples collected from different patients in Thailand. The test sample is added to the sample well of the test cassette, where AuNP-RBD-IgY conjugates react with human anti-SARS-CoV-2 IgG and IgM in the sample. The sample then migrates through capillary action through the test and control lines. Sample #2 was found to be negative for both IgM and IgG, #86 was positive for IgM and IgG, while #32 was positive for IgM only. AuNPs: gold nanoparticles; C: control line; G: test line for human IgG; M: test line for human IgM; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
Discussion
The serological testing of COVID-19 is crucial for disease management, and will help identify asymptomatic cases that could spread the virus further. As current vaccines or therapeutic measures are inadequate to control or treat the disease, prevention is the only option available to check and prevent the spread of infection.22, 23 Current control measures aim at reducing the outbreaks of COVID-19. However, accurate, early diagnosis of the disease is critical to minimize further spread of the virus and resultant human losses. Consequently there is an urgent need to detect acute and chronic diseases in a simple, affordable, and rapid manner, which was the primary objective of the present study. Hence, an LFIA was developed using a recombinant plant-produced RBD antigen coupled with AuNPs as the detection label.
Plant expression systems offer many advantages for the production of recombinant proteins, which can be used as diagnostic reagents, vaccines, and drugs.24 The main benefits include cost-effectiveness, the speed of transient expression for rapid large-scale production of recombinant proteins, and the system’s safety as plants do not host any known human pathogens.25 The plant-purified antigen was further used to make an LFIA cassette. The developed LFIA kit is intended for the in vitro qualitative detection of IgG and IgM antibodies against SARS-CoV-2 in human samples. The test results can be obtained in less than 15 minutes, and the results assessed visually. The LFIA developed here could make a considerable economic impact in reducing the costs of testing and reducing human mortality by identifying asymptomatic cases and containing the further spread of the virus. Usually, healthcare workers in hospitals and laboratories, especially in developing and under-developed countries, have to screen samples for COVID-19 mainly by polymerase chain reaction testing, which incurs a high cost. In contrast, the LFIA is comparatively cost-effective, can be performed even without professional help and could serve as a preliminary screening test to screen susceptible samples.26, 27 Thus, the presented study demonstrates the potential of a plant expression system for the rapid large-scale production of recombinant proteins during epidemic situations and the efficiency of LFIA technology for achieving rapid non-laborious point-of-care diagnosis of COVID-19. To the best of our knowledge, this is the first diagnostic kit that uses a plant-produced recombinant protein against SARS-CoV-2 for the diagnosis of COVID-19. The developed assay could represent a considerable improvement in COVID-19 diagnosis.
In summary, we evaluated a rapid point-of-care COVID-19-specific LFIA developed using a plant-produced recombinant RBD antigen to detect SARS-CoV-2-specific IgG and IgM. The specific recognition of RBD-SARS-CoV-2 by the IgG/IgM antibodies of COVID-19-infected patients emphasizes the potential of the plant-produced RBD protein for COVID-19 serodiagnosis. The developed LFIA device will help healthcare workers detect the presence of antibodies in patient serum samples in less than 15 minutes. This rapid screening test offers many advantages, including a reduced requirement for sample processing requirement, cost-effectiveness, and ease of use, as well as being easily portable, time-efficient, and technologically feasible, making these assays compatible with clinical application in Thai hospitals. Because patients with a post-dengue virus infection or with rheumatoid factor IgM possibly produced false results of LFIA.28, 29 Baiya’ Rapid COVID-19’ rapid IgM/IgG test kit should be the further test to identify these interferences from various patient conditions.
Author contributions
KR, GY, BS and WP performed the expression and analysis of SARS-CoV-RBD protein. KK, PS, EP and ST performed the analysis and collected the data. ST and WP conceived and designed the analysis and managed the research project. All authors wrote, revised, and approved the manuscript.
Financial support
This study was funded by Baiya Phytopharm Co., Ltd.
Acknowledgement
The authors would like to thank Baiya Phytopharm Co., Ltd. for providing the financial support.
Conflicts of interest statement
Suthira Taychakhoonavudh and Waranyoo Phoolcharoen from Chulalongkorn University are co-founders/shareholders of Baiya Phytopharm Co., Ltd.
Data sharing statement
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Reference
Statement on the second meeting of the international health regulations emergency committee regarding the outbreak of novel coronavirus (2019-nCoV)
Diagnostic testing for severe acute respiratory syndrome-related coronavirus 2: a narrative review
DOI:10.7326/M20-1301
URL
PMID:32282894
[Cited within: 1]

Diagnostic testing to identify persons infected with severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) infection is central to control the global pandemic of COVID-19 that began in late 2019. In a few countries, the use of diagnostic testing on a massive scale has been a cornerstone of successful containment strategies. In contrast, the United States, hampered by limited testing capacity, has prioritized testing for specific groups of persons. Real-time reverse transcriptase polymerase chain reaction-based assays performed in a laboratory on respiratory specimens are the reference standard for COVID-19 diagnostics. However, point-of-care technologies and serologic immunoassays are rapidly emerging. Although excellent tools exist for the diagnosis of symptomatic patients in well-equipped laboratories, important gaps remain in screening asymptomatic persons in the incubation phase, as well as in the accurate determination of live viral shedding during convalescence to inform decisions to end isolation. Many affluent countries have encountered challenges in test delivery and specimen collection that have inhibited rapid increases in testing capacity. These challenges may be even greater in low-resource settings. Urgent clinical and public health needs currently drive an unprecedented global effort to increase testing capacity for SARS-CoV-2 infection. Here, the authors review the current array of tests for SARS-CoV-2, highlight gaps in current diagnostic capacity, and propose potential solutions.
COVID-19: Progress in diagnostics, therapy and vaccination
DOI:10.7150/thno.47987
URL
PMID:32685022
[Cited within: 1]

Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has recently become a pandemic. As the sudden emergence and rapid spread of SARS-CoV-2 is endangering global health and the economy, the development of strategies to contain the virus's spread are urgently needed. At present, various diagnostic kits to test for SARS-CoV-2 are available for use to initiate appropriate treatment faster and to limit further spread of the virus. Several drugs have demonstrated in vitro activity against SARS-CoV-2 or potential clinical benefits. In addition, institutions and companies worldwide are working tirelessly to develop treatments and vaccines against COVID-19. However, no drug or vaccine has yet been specifically approved for COVID-19. Given the urgency of the outbreak, we focus here on recent advances in the diagnostics, treatment, and vaccine development for SARS-CoV-2 infection, helping to guide strategies to address the current COVID-19 pandemic.
Laboratory diagnosis of emerging human coronavirus infections - the state of the art
DOI:10.1080/22221751.2020.1745095
URL
PMID:32196430
[Cited within: 1]

The three unprecedented outbreaks of emerging human coronavirus (HCoV) infections at the beginning of the twenty-first century have highlighted the necessity for readily available, accurate and fast diagnostic testing methods. The laboratory diagnostic methods for human coronavirus infections have evolved substantially, with the development of novel assays as well as the availability of updated tests for emerging ones. Newer laboratory methods are fast, highly sensitive and specific, and are gradually replacing the conventional gold standards. This presentation reviews the current laboratory methods available for testing coronaviruses by focusing on the coronavirus disease 2019 (COVID-19) outbreak going on in Wuhan. Viral pneumonias typically do not result in the production of purulent sputum. Thus, a nasopharyngeal swab is usually the collection method used to obtain a specimen for testing. Nasopharyngeal specimens may miss some infections; a deeper specimen may need to be obtained by bronchoscopy. Alternatively, repeated testing can be used because over time, the likelihood of the SARS-CoV-2 being present in the nasopharynx increases. Several integrated, random-access, point-of-care molecular devices are currently under development for fast and accurate diagnosis of SARS-CoV-2 infections. These assays are simple, fast and safe and can be used in the local hospitals and clinics bearing the burden of identifying and treating patients.
Lateral Flow immunoassays for Ebola virus disease detection in liberia
DOI:10.1093/infdis/jiw251
URL
PMID:27443616
[Cited within: 1]

BACKGROUND: Lateral flow immunoassays (LFIs) are point-of-care diagnostic assays that are designed for single use outside a formal laboratory, with in-home pregnancy tests the best-known example of these tests. Although the LFI has some limitations over more-complex immunoassay procedures, such as reduced sensitivity and the potential for false-positive results when using complex sample matrices, the assay has the benefits of a rapid time to result and ease of use. These benefits make it an attractive option for obtaining rapid results in an austere environment. In an outbreak of any magnitude, a field-based rapid diagnostic assay would allow proper patient transport and for safe burials to be conducted without the delay caused by transport of samples between remote villages and testing facilities. Use of such point-of-care instruments in the ongoing Ebola virus disease (EVD) outbreak in West Africa would have distinct advantages in control and prevention of local outbreaks, but proper understanding of the technology and interpretation of results are important. METHODS: In this study, a LFI, originally developed by the Naval Medical Research Center for Ebola virus environmental testing, was evaluated for its ability to detect the virus in clinical samples in Liberia. Clinical blood and plasma samples and post mortem oral swabs submitted to the Liberian Institute for Biomedical Research, the National Public Health Reference Laboratory for EVD testing, were tested and compared to results of real-time reverse transcription-polymerase chain reaction (rRT-PCR), using assays targeting Ebola virus glycoprotein and nucleoprotein. RESULTS: The LFI findings correlated well with those of the real-time RT-PCR assays used as benchmarks. CONCLUSIONS: Rapid antigen-detection tests such as LFIs are attractive alternatives to traditional immunoassays but have reduced sensitivity and specificity, resulting in increases in false-positive and false-negative results. An understanding of the strengths, weaknesses, and limitations of a particular assay lets the diagnostician choose the correct situation to use the correct assay and properly interpret the results.
COVID-19 serological tests: how well do they actually perform?
Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies
DOI:10.1016/j.jcv.2020.104413
URL
PMID:32403010
[Cited within: 1]

INTRODUCTION: Several SARS-CoV-2 immunoassays have been developed recently. The purpose of this study was to assess the performance of five immunoassays for the detection of SARS-CoV-2 antibodies. METHODS: Two quantitative automated immunoassays (Maglumi2019-n-Cov IgG and IgM and Euroimmun Anti-SARS-CoV-2 IgG and IgA assays) and three lateral flow rapid tests were performed. This retrospective study included 200 residual sera from patients and healthy volunteers. Case serum samples (n = 128) were obtained from COVID-19 patients confirmed by RT-qPCR and CT-scan. Days since onset of symptoms was collected from their medical records. Control non-SARS-CoV-2 samples (n = 72) were obtained from anonymous stored residual serum samples. RESULTS: Maglumi IgG/IgM tests showed overall less sensitivity than Euroimmun IgG/IgA test (84.4 % versus 64.3 %). Both tests showed similar specificities of IgG at 99 % and 100 %, respectively. The results from the lateral flow assays were easily interpretable with unambiguous coloured reading bands. The overall sensitivity of the three tests was similar (around 70 %) without any significant differences. The sensitivity of the three lateral flow assays and also of the serological quantitative assays increased during the second week after symptom onset and all reached similar values (91 %-94 %) after 14 days. CONCLUSION: This study shows accurate and equivalent performance of the five serological antibody assays (ELISA, CLIA and three lateral flow tests) in detecting SARS-CoV-2 antibodies 14 days after the onset of COVID-19 symptoms. This is compatible with their application in specific clinical contexts and in determining epidemiological strategies for the COVID-19 pandemic.
Perspectives on monoclonal antibody therapy as potential therapeutic intervention for coronavirus disease-19 (COVID-19)
DOI:10.12932/AP-200220-0773
URL
PMID:32134278
[Cited within: 1]

Last decade witnessed the outbreak of many life-threatening human pathogens including Nipah, Ebola, Chikungunya, Zika, Middle East respiratory syndrome coronavirus (MERS-CoV), Severe Acute respiratory syndrome coronavirus (SARS-CoV) and more recently novel coronavirus (2019-nCoV or SARS-CoV-2). The disease condition associated with novel coronavirus, referred to as Coronavirus disease (COVID-19). The emergence of novel coronavirus in 2019 in Wuhan, China marked the third highly pathogenic coronavirus infecting humans in the 21st century. The continuing emergence of coronaviruses at regular intervals poses a significant threat to human health and economy. Ironically, even after a decade of research on coronavirus, still there are no licensed vaccines or therapeutic agents to treat coronavirus infection which highlights an urgent need to develop effective vaccines or post-exposure prophylaxis to prevent future epidemics. Several clinical, genetic and epidemiological features of COVID-19 resemble SARS-CoV infection. Hence, the research advancements on SARS-CoV treatment might help scientific community in quick understanding of this virus pathogenesis and develop effective therapeutic/prophylactic agents to treat and prevent this infection. Monoclonal antibodies represent the major class of biotherapeutics for passive immunotherapy to fight against viral infection. The therapeutic potential of monoclonal antibodies has been well recognized in the treatment of many diseases. Here, we summarize the potential monoclonal antibody based therapeutic intervention for COVID-19 by considering the existing knowledge on the neutralizing monoclonal antibodies against similar coronaviruses SARS-CoV and MERS-CoV. Further research on COVID-19 pathogenesis could identify appropriate therapeutic targets to develop specific anti-virals against this newly emerging pathogen.
A promising vaccine candidate against COVID-19
Plant-made pharmaceuticals: leading products and production platforms
DOI:10.1002/bab.6
URL
PMID:21446960
[Cited within: 1]

The number of approaches to recombinant protein production in plants is greater than ever before. Development of these new and improved technologies as production platforms for plant-made pharmaceuticals has and will continue to create new commercial opportunities in the pharmaceutical sector. However, it is inevitable that no single system will be optimal for the production of all recombinant proteins of interest in plants due to both the physical characteristics and the envisaged therapeutic application of each product. Here, we review a range of promising product/platform pairs emphasizing synergies during production and in clinical trials.
Expression of an immunogenic Ebola immune complex in Nicotiana benthamiana
DOI:10.1111/j.1467-7652.2011.00593.x
URL
PMID:21281425

Filoviruses (Ebola and Marburg viruses) cause severe and often fatal haemorrhagic fever in humans and non-human primates. The US Centers for Disease Control identifies Ebola and Marburg viruses as 'category A' pathogens (defined as posing a risk to national security as bioterrorism agents), which has lead to a search for vaccines that could prevent the disease. Because the use of such vaccines would be in the service of public health, the cost of production is an important component of their development. The use of plant biotechnology is one possible way to cost-effectively produce subunit vaccines. In this work, a geminiviral replicon system was used to produce an Ebola immune complex (EIC) in Nicotiana benthamiana. Ebola glycoprotein (GP1) was fused at the C-terminus of the heavy chain of humanized 6D8 IgG monoclonal antibody, which specifically binds to a linear epitope on GP1. Co-expression of the GP1-heavy chain fusion and the 6D8 light chain using a geminiviral vector in leaves of N. benthamiana produced assembled immunoglobulin, which was purified by ammonium sulphate precipitation and protein G affinity chromatography. Immune complex formation was confirmed by assays to show that the recombinant protein bound the complement factor C1q. Size measurements of purified recombinant protein by dynamic light scattering and size-exclusion chromatography also indicated complex formation. Subcutaneous immunization of BALB/C mice with purified EIC resulted in anti-Ebola virus antibody production at levels comparable to those obtained with a GP1 virus-like particle. These results show excellent potential for a plant-expressed EIC as a human vaccine.
A novel system for rapid and cost-effective production of detection and diagnostic reagents of West Nile virus in plants
DOI:10.1155/2012/106783
URL
PMID:22187532

The threat of West Nile virus (WNV) epidemics necessitates the development of a technology platform that can produce reagents to support detection and diagnosis rapidly and inexpensively. A plant expression system is attractive for protein production due to its low-cost and high-scalability nature and its ability to make appropriate posttranslational modifications. Here, we investigated the feasibility of using plants to produce two WNV detection and diagnostic reagents to address the current cost and scalability issues. We demonstrated that WNV DIII antigen and E16 monoclonal antibody are rapidly produced at high levels in two plant species and are easily purified. Furthermore, they are effective in identifying WNV and in detecting human IgM response to WNV infection. E16 mAb does not cross-react with other flaviviruses, therefore, is valuable for improving diagnostic accuracy. This study provides a proof of principle for using plants as a robust and economical system to produce diagnostic reagents for arboviruses.
Plant expression platform for the production of recombinant pharmaceutical proteins
Plant-produced candidate countermeasures against emerging and reemerging infections and bioterror agents
DOI:10.1111/pbi.12475
URL
PMID:26387510

Despite progress in the prevention and treatment of infectious diseases, they continue to present a major threat to public health. The frequency of emerging and reemerging infections and the risk of bioterrorism warrant significant efforts towards the development of prophylactic and therapeutic countermeasures. Vaccines are the mainstay of infectious disease prophylaxis. Traditional vaccines, however, are failing to satisfy the global demand because of limited scalability of production systems, long production timelines and product safety concerns. Subunit vaccines are a highly promising alternative to traditional vaccines. Subunit vaccines, as well as monoclonal antibodies and other therapeutic proteins, can be produced in heterologous expression systems based on bacteria, yeast, insect cells or mammalian cells, in shorter times and at higher quantities, and are efficacious and safe. However, current recombinant systems have certain limitations associated with production capacity and cost. Plants are emerging as a promising platform for recombinant protein production due to time and cost efficiency, scalability, lack of harboured mammalian pathogens and possession of the machinery for eukaryotic post-translational protein modification. So far, a variety of subunit vaccines, monoclonal antibodies and therapeutic proteins (antivirals) have been produced in plants as candidate countermeasures against emerging, reemerging and bioterrorism-related infections. Many of these have been extensively evaluated in animal models and some have shown safety and immunogenicity in clinical trials. Here, we overview ongoing efforts to producing such plant-based countermeasures.
Development of systems for the production of plant-derived biopharmaceuticals
Transient expression of dengue virus NS1 antigen in Nicotiana benthamiana for use as a diagnostic antigen
DOI:10.3389/fpls.2019.01674
URL
PMID:32010161

Dengue is a viral disease that represents a significant threat to global public health since billions of people are now at risk of infection by this mosquito-borne virus. The implementation of extensive screening tests is indispensable to control this disease, and the Dengue virus non-structural protein 1 (NS1) is a promising antigen for the serological diagnosis of dengue fever. Plant-based systems can be a safe and cost-effective alternative for the production of dengue virus antigens. In this work, two strategies to produce the dengue NS1 protein in Nicotiana benthamiana leaves were evaluated: Targeting NS1 to five different subcellular compartments to assess the best subcellular organelle for the expression and accumulation of NS1, and the addition of elastin-like polypeptide (ELP) or hydrophobin (HFBI) fusion tags to NS1. The transiently expressed proteins in N. benthamiana were quantified by Western blot analysis. The NS1 fused to ELP and targeted to the ER (NS1 ELP-ER) showed the highest yield (445 mg/kg), approximately a forty-fold increase in accumulation levels compared to the non-fused protein (NS1-ER), representing the first example of transient expression of DENV NS1 in plant. We also demonstrated that NS1 ELP-ER was successfully recognized by a monoclonal anti-dengue virus NS1 glycoprotein antibody, and by sera from dengue virus-infected patients. Interestingly, it was found that transient production of NS1-ER and NS1 ELP-ER using vacuum infiltration of whole plants, which is easier to scale up, rather than syringe infiltration of leaves, greatly improved the accumulation of NS1 proteins. The generated plant made NS1, even without extensive purification, showed potential to be used for the development of the NS1 diagnostic tests in resource-limited areas where dengue is endemic.
Plant molecular farming: a viable platform for recombinant biopharmaceutical production
Rapid production of SARS-CoV-2 receptor binding domain (RBD) and spike specific monoclonal antibody CR3022 in Nicotiana benthamiana
DOI:10.1038/s41598-020-74904-1
URL
PMID:33077899
[Cited within: 2]

Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is responsible for the ongoing global outbreak of coronavirus disease (COVID-19) which is a significant threat to global public health. The rapid spread of COVID-19 necessitates the development of cost-effective technology platforms for the production of vaccines, drugs, and protein reagents for appropriate disease diagnosis and treatment. In this study, we explored the possibility of producing the receptor binding domain (RBD) of SARS-CoV-2 and an anti-SARS-CoV monoclonal antibody (mAb) CR3022 in Nicotiana benthamiana. Both RBD and mAb CR3022 were transiently produced with the highest expression level of 8 mug/g and 130 mug/g leaf fresh weight respectively at 3 days post-infiltration. The plant-produced RBD exhibited specific binding to the SARS-CoV-2 receptor, angiotensin-converting enzyme 2 (ACE2). Furthermore, the plant-produced mAb CR3022 binds to SARS-CoV-2, but fails to neutralize the virus in vitro. This is the first report showing the production of anti-SARS-CoV-2 RBD and mAb CR3022 in plants. Overall these findings provide a proof-of-concept for using plants as an expression system for the production of SARS-CoV-2 antigens and antibodies or similar other diagnostic reagents against SARS-CoV-2 rapidly, especially during epidemic or pandemic situation.
Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay
DOI:10.1021/acsomega.0c01554
URL
PMID:32542208
[Cited within: 1]

Last year, the novel coronavirus disease (COVID-19) emerged in Wuhan, and it has rapidly spread to many other countries and regions. COVID-19 exhibits a strong human-to-human transmission infectivity and could cause acute respiratory diseases. Asymptomatic carriers are able to infect other healthy persons, and this poses a challenge for public health; the World Health Organization (WHO) has already announced COVID-19 as a global pandemic. Nucleic acid testing, considered as the current primary method for diagnosing COVID-19, might lead to false negatives and is difficult to be applied for every suspected patient because of the existence of asymptomatic carriers. Meanwhile, detecting specific antibodies in blood, such as the IgM antibody, against the SARS-CoV-2 virus is another choice for COVID-19 diagnosis, as it is widely accepted that IgM is an important indicator in the acute infection period. In this study, a colloidal gold nanoparticle-based lateral-flow (AuNP-LF) assay was developed to achieve rapid diagnosis and on-site detection of the IgM antibody against the SARS-CoV-2 virus through the indirect immunochromatography method. For preparing AuNP-LF strips, the SARS-CoV-2 nucleoprotein (SARS-CoV-2 NP) was coated on an analytical membrane for sample capture, and antihuman IgM was conjugated with AuNPs to form the detecting reporter. Optimization of AuNP-LF assay was carried out by altering the pH value and the amount of antihuman IgM. The performance of AuNP-LF assay was evaluated by testing serum samples of COVID-19 patients and normal humans. The results were compared with the real-time polymerase chain reaction. The sensitivity and specificity of AuNP-LF assay were determined to be 100 and 93.3%, respectively, and an almost perfect agreement was exhibited by Kappa statistics (kappa coefficient = 0.872). AuNP-LF assay showed outstanding selectivity in the detection of IgM against the SARS-CoV-2 virus with no interference from other viruses such as severe fever with thrombocytopenia syndrome virus (SFTSV) and dengue virus (DFV). AuNP-LF assay was able to achieve results within 15 min and needed only 10-20 muL serum for each test. As a whole, in the light of its advantages such as excellent specificity and stability, easy operation, low cost, and being less time-consuming, AuNP-LF assay is a feasible method for the diagnosis of COVID-19 in primary hospitals and laboratories, especially in emergency situations in which numerous samples need to be tested on time.
A multi-target lateral flow immunoassay enabling the specific and sensitive detection of total antibodies to SARS COV-2
DOI:10.1016/j.talanta.2020.121737
URL
PMID:33303174
[Cited within: 1]

A rapid test for detecting total immunoglobulins directed towards the nucleocapsid protein (N) of severe acute syndrome coronavirus 2 (SARS CoV-2) was developed, based on a multi-target lateral flow immunoassay comprising two test lines. Both test lines bound to several classes of immunoglobulins (G, M, and A). Specific anti-SARS immunoglobulins were revealed by a colorimetric probe formed by N and gold nanoparticles. Targeting the total antibodies response to infection enabled achieving 100% diagnostic specificity (95.75-100, C.I. 95%, n = 85 healthy and with other infections individuals) and 94.6% sensitivity (84.9-98.9, C.I. 95%, n = 62 SARS CoV-2 infected subjects) as early as 7 days post confirmation of positivity. Agreeing results with a reference serological ELISA were achieved, except for the earlier detection capability of the rapid test. Follow up of the three seroconverting patients endorsed the hypothesis of the random rise of the different immunoglobulins and strengthened the 'total antibodies' approach for the trustworthy detection of serological response to SARS CoV-2 infection.
Emergence of novel coronavirus 2019-nCoV: need for rapid vaccine and biologics development
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an emerging zoonotic respiratory pathogen in humans
Plant molecular farming - integration and exploitation of side streams to achieve sustainable biomanufacturing
DOI:10.3389/fpls.2018.01893
URL
PMID:30713542
[Cited within: 1]

Plants have unique advantages over other systems such as mammalian cells for the production of valuable small molecules and proteins. The benefits cited most often include safety due to the absence of replicating human pathogens, simplicity because sterility is not required during production, scalability due to the potential for open-field cultivation with transgenic plants, and the speed of transient expression potentially providing gram quantities of product in less than 4 weeks. Initially there were also significant drawbacks, such as the need to clarify feed streams with a high particle burden and the large quantities of host cell proteins, but efficient clarification is now readily achieved. Several additional advantages have also emerged reflecting the fact that plants are essentially biodegradable, single-use bioreactors. This article will focus on the exploitation of this concept for the production of biopharmaceutical proteins, thus improving overall process economics. Specifically, we will discuss the single-use properties of plants, the sustainability of the production platform, and the commercial potential of different biomass side streams. We find that incorporating these side streams through rational process integration has the potential to more than double the revenue that can currently be achieved using plant-based production systems.
Advances in plant molecular farming
Assay techniques and test development for COVID-19 diagnosis
DOI:10.1021/acscentsci.0c00501 URL PMID:32382657 [Cited within: 1]
In vitro diagnostic assays for COVID-19: recent advances and emerging trends
Potential antigenic cross-reactivity between SARS-CoV-2 and Dengue viruses
DOI:10.1093/cid/ciab257 URL PMID:33754646 [Cited within: 1]
A method to prevent SARS-CoV-2 IgM false positives in gold immunochromatography and enzyme-linked immunosorbent assays