Introduction
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late 2019 and has spread widely across the world now.1 As of December 21, 2020, SARS-CoV-2 had infected 75,704,857 people and resulted in 1,690,061 deaths worldwide.2 SARS-CoV-2 is considered the causative agent of coronavirus disease 2019 (COVID-19), a respiratory disease characterized by a range of symptoms—or lack thereof—that vary with age and pre-existing health conditions, which can lead to hospitalization and strain the healthcare system.1 Despite improvements in treatment and public policy aimed to curb the spread of the virus, cases remain high and have been rapidly increasing since November, 2020 in many regions across the world.
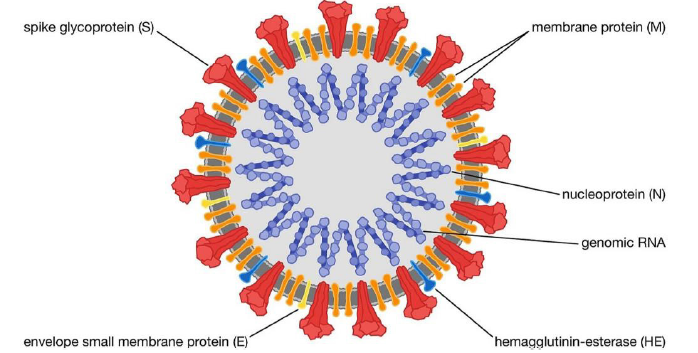
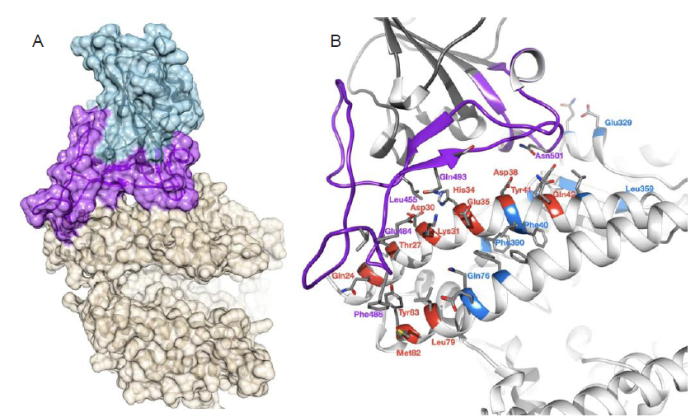
Like the previous human coronaviruses severe acute respiratory syndrome-associated coronavirus (SARS-CoV) and Middle East respiratory syndrome (MERS), SARS-CoV-2 is a betacoronavirus that is likely of zoonotic origin, as suggested by its genetic similarity with betacoronaviruses found in bats and pangolins.1 SARS-CoV-2 contains single-stranded RNA that is surrounded by a protein envelope, which contains crown-like spike proteins on the outer surface.3 Structurally, SARS-CoV-2 is composed of four structural proteins, namely spike (S), envelope (E), membrane (M), and nucleocapsid (N), as well as the replicase open reading frame (ORF1a/ORF1b), which encodes a polypeptide that is cleaved to form assorted non-structural proteins involved in replication and transcription (Figure 1).1, 4 Of interest is the S protein, which mediates the entry of SARS-CoV-2 into host cells. The viral S protein is composed of two subunits, designated S1 and S2, of which the former contains the N-terminal domain and the receptor-binding domain (RBD), while the latter contains the fusion peptide, transmembrane domain, cytoplasmic domain, and two heptapeptide repeat sequences (HR1 and HR2).5 These two subunits are responsible for recognizing and binding to host angiotensin converting enzyme II receptors and subsequent cell fusion (Figure 2).5, 6
Figure 1.
Figure 1.
Structure of SARS-CoV-2. A graphic illustrating the structure of SARS-CoV-2, which shows the viral RNA along with the S, M, E, and N proteins. Figure reprinted from Shaikh et al.4 Licensed under CC BY 4.0. SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
Figure 2.
Figure 2.
The SARS-CoV-2 spike protein bound to the ACE2 receptor. (A) The spike protein RBD (light blue, purple) is shown containing the receptor-binding motif (purple) while at the interface of the ACE2 receptor (tan). (B) Interface residues of the RBD (purple) are shown interacting with ACE2 residues in direct contact (red) or extended direct contact (blue) with the RBD. Figure reprinted from Lam et al.6 Licensed under CC BY 4.0. ACE2: angiotensin converting enzyme II; RBD: receptor-binding domain; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
Angiotensin converting enzyme II receptors are commonly expressed by epithelial alveolar type II cells in the lungs, as well as in the heart, kidneys, and intestines.7 Viral cell fusion is mediated by a number of host furin-like proteases, such as trypsin and transmembrane serine protease 2, which cleave the S protein into S1 and S2 subunits at furin cleavage sites. It is thought that the greater number of furin cleavage sites in the S protein of SARS-CoV-2 is responsible for its greater pathogenicity compared to SARS-CoV. After cleavage of the S protein, the fusion peptide binds to the host cell membrane and initiates fusion. The HR1 and HR2 domains then bring the two membranes together until they fuse, allowing the virus to release genetic material into the host cell.5 The role of the S protein in SARS-CoV-2 pathogenicity makes it a key target for vaccine development, though E, M, N, and other accessory proteins may also hold potential to act as antigens.
Development of a safe and effective vaccine requires an understanding of the immune correlates of protection against SARS-CoV-2. It has been found that infection with SARS-CoV-2 induces both humoral and cellular immune responses.1 The production of neutralizing antibodies seems to provide a good correlate of protection against SARS-CoV-2. A study using purified IgG antibodies from convalescent rhesus macaques was found to confer protection for rhesus macaques with no previous exposure to the virus, which seems to indicate their role in protecting against SARS-CoV-2 reinfection.8 That study also found that CD8+ T cells can mediate protection against SARS-CoV-2 reinfection in the wake of waning antibody titers.8 A follow up with SARS-CoV patients six years post infection identified memory T cell responses, even with no detectable IgG antibodies or memory B cell responses.9 These results may imply the potential for a long-lasting cellular response to SARS-CoV-2 even after waning antibody titres. Thus, vaccine candidates should induce both humoral and cellular responses against SARS-CoV-2.
In response to the COVID-19 pandemic, vaccines are being developed at an unprecedented speed using various novel materials and technologies representing the most advanced biomedical science. Countries, seeking to mitigate economic disruptions and loss of life, are committing to mass vaccination programs as soon as a vaccine candidate is deemed safe and effective. The goal of these programs is to quickly reach a state of herd immunity, which would likely require unnecessary loss of life and economic productivity if left to occur naturally, as in the 1918 Spanish influenza pandemic.10 Thus, safe and effective vaccines seem to be the best method of ending the COVID-19 pandemic. In this paper we are going to summarize the development and mechanisms of several of the novel vaccine types that have been developed for SARS-CoV-2.
The articles used in this review of the COVID-19 vaccines were retrieved through an electronic search of the PubMed database. Literatures from 2019 to present with regards to COVID-19 and the COVID-19 vaccines were included. Initial searches were performed under the following conditions: ((COVID-19) OR (SARS-2-CoV)) AND (vaccines). Studies were screened by title, abstract, and date to include only human COVID-19 vaccines, as well as the most up-to-date studies. Subsequent searches were completed relevant to the different types of COVID-19 vaccine using the following terms: RNA-based vaccine, DNA-based vaccine, protein subunit vaccine, recombinant protein vaccine, viral vector vaccine, adenovirus vector vaccine, adjuvants, and cold-chain transport.
Different Types of COVID-19 Vaccine
According to the World Health Organization, there are 64 vaccine candidates already in clinical trials and 172 candidates in pre-clinical development as of January 6, 2021.2 Table 1 lists the vaccines currently undergoing clinical trials, as well as those authorized for limited or emergency use in certain regions. Of the vaccine mechanisms to be discussed, 30% utilize protein subunits, while 14% use an inactivated virus and 27% use a viral vector, either replicating or non-replicating. Additionally, of the 24% of vaccine candidates that use nucleic acids, 13% are DNA-based while 11% are RNA-based.2
Table 1 Summary of COVID-19 vaccines currently in clinical trials
| Vaccine candidate | Company | Mechanism | Phase |
|---|---|---|---|
| SARS-CoV-2 vaccine | Sinovac Research and Development Co., Ltd. | Inactivated | Phase 3 |
| Inactivated SARS-CoV-2 vaccine | Sinopharm + China National Biotec Group Co. Ltd. + Wuhan Institute of Biological Products | Inactivated | Phase 3 |
| Inactivated SARS-CoV-2 vaccine | Sinopharm + China National Biotec Group Co. Ltd. + Beijing Institute of Biological Products | Inactivated | Phase 3 |
| ChAdOx1-S (AZD1222) | AstraZeneca + University of Oxford | Viral vector | Phase 3 |
| Recombinant novel coronavirus vaccine (adenovirus type 5 vector) | CanSino Biologics Inc. + Beijing Institute of Biotechnology | Viral vector | Phase 3 |
| Gam-COVID-Vac, Aden-based (rAd26-S+rAd5-S) | Gamaleya Research Institute, Health Ministry of the Russian Federation | Viral vector | Phase 3 |
| AD26.COV2.S | Janssen Pharmaceuticals, Inc. | Viral vector | Phase 3 |
| SARS-CoV-2 rS/Matrix M1-Adjuvant | Novavax | Protein subunit | Phase 3 |
| mRNA-1273 | Moderna + National Institute of Allergy and Infectious Diseases | RNA | Phase 3 |
| BNT162 (3 LNP-mRNAs) | BioNTech + Fosun Pharma; Jiangsu Provincial Centre for Disease Prevention and Control + Pfizer | RNA | Phase 2/3 |
| Recombinant SARS-CoV-2 vaccine | Anhui Zhifei Longcom Biopharmaceuticals + Institute of Microbiology, Chinese Academy of Sciences | Protein subunit | Phase 3 |
| CVnCoV vaccine | CureVac AG | RNA | Phase 3 |
| SARS-CoV-2 vaccine | Institute of Medical Biology, Chinese Academy of Medical Sciences | Inactivated | Phase 3 |
| QazCovid-in - COVID-19 inactivated vaccine | Research Institute for Biological Safety Problems, Republic of Kazakhstan | Inactivated | Phase 3 |
| INO-4800+electroporation | Inovio Pharmaceuticals + International Vaccine Institute, South Korea + Advaccine (Suzhou) Biopharmaceutical Co., Ltd. | DNA | Phase 2/3 |
| AG0301-COVID19 | AnGes + Takara Bio Inc. + Osaka University | DNA | Phase 2/3 |
| nCov vaccine | Cadila Healthcare Ltd. | DNA | Phase 3 |
| GX-19 | Genexine Consortium | DNA | Phase 1/2 |
| Whole-Virion Inactivated SARS-CoV-2 Vaccine (BBV152) | Bharat Biotech International Limited | Inactivated | Phase 3 |
| KBP-COVID-19 (RBD-based) | Kentucky Bioprocessing Inc. | Protein subunit | Phase 1/2 |
| SARS-CoV-2 vaccine formulation 1 with adjuvant | Sanofi Pasteur + GSK | Protein subunit | Phase 1/2 |
| ARCT-021 | Arcturus Therapeutics | RNA | Phase 2 |
| RBD SARS-CoV-2 HBsAg VLP vaccine | Serum Institute of India + Accelagen Pty | Virus like particle | Phase 1/2 |
| Inactivated SARS-CoV-2 vaccine | Shenzhen Kangtai Biological Products Co., Ltd. | Inactivated | Phase 2 |
| GRAd-COV2 | ReiThera + Leukocare + Univercells | Viral vector | Phase 1 |
| VXA-CoV2-1 AD5 adjuvanted oral vaccine platform | Vaxart Inc. | Viral vector | Phase 1 |
| MVA-SARS-2-S | University Medical Centre Hamburg-Eppendorf + Ludwig Maximilian University of Munich | Viral vector | Phase 2 |
| SCB-2019 + AS03 or CpG 1018 adjuvant plus Alum adjuvant | Clover Biopharmaceuticals Inc./GSK/Dynavax | Protein subunit | Phase 2/3 |
| COVID19 vaccine | Vaxine Pty Ltd. + Medytox | Protein subunit | Phase 1 |
| MVC-COV1901 (S-2P protein + CpG 1018) | Medigen Vaccine Biologics + Dynavax + National Institute of Allergy and Infectious Diseases | Protein subunit | Phase 1 |
| FINLAY-FR anti-SARS-CoV-2 Vaccine | Instituto Finlay de Vacunas | Protein subunit | Phase 2 |
| EpiVacCorona | Federal Budgetary Research Institution, State Research Centre of Virology and Biotechnology “Vector” | Protein subunit | Phase 1/2 |
| RBD Recombinant SARS-CoV-2 vaccine (Sf9 cell) | West China Hospital of Sichuan University | Protein subunit | Phase 2 |
| IMP CoVac-1 (SARS-CoV-2 HLA-DR peptides) | University Hospital Tübingen | Protein subunit | Phase 1 |
| UB-612 | COVAXX + United Biomedical Inc. | Protein subunit | Phase 2/3 |
| V591-001 - Measles-vector based (TMV-o38) | Merck & Co. Inc. + Themis + Merck Sharp & Dohme Ltd. + Institut Pasteur + University of Pittsburgh | Viral vector (replicating) | Phase 1/2 |
| DelNS1-2019-nCoV-RBD-OPT1 | Jiangsu Provincial Centre for Disease Prevention and Control | Viral vector (replicating) | Phase 2 |
| LNP-nCoVsaRNA | Imperial College London | RNA | Phase 1 |
| SARS-CoV-2 mRNA vaccine | Shulan Hospital + Guangxi Centre for Disease Prevention and Control | RNA | Phase 1 |
| Coronavirus-like particle COVID-19 | Medicago Inc. | Viral like particle | Phase 2/3 |
| Covid-19/aAPC vaccine | Shenzhen Geno-Immune Medical Institute | Viral vector (replicating) + APC | Phase 1 |
| LV-SMENP-DC vaccine | Shenzhen Geno-Immune Medical Institute | Viral vector (non-replicating) + APC | Phase 1/2 |
| AdimrSC-2f | Adimmune Corporation | Protein subunit | Phase 1 |
| Covigenix VAX-001 | Entos Pharmaceuticals Inc. | DNA | Phase 1 |
| CORVax | Providence Health & Services | DNA | Phase 1 |
| ChulaCov19 mRNA vaccine | Chulalongkorn University | RNA | Phase 1 |
| bacTRL-Spike | Symvivo Corporation | DNA | Phase 1 |
| hAd5-S-Fusion+N-ETSD vaccine | ImmunityBio, Inc. | Viral vector | Phase 1 |
| COH04S1 (MVA-SARS-2-S) | City of Hope Medical Center + National Cancer Institute | Viral vector | Phase 1 |
| rVSV-SARS-CoV-2-S vaccine | Israel Institute for Biological Research | Viral vector (replicating) | Phase 1/2 |
| Dendritic cell vaccine AV-COVID-19 | Avita Biomedical, Inc. + National Institute of Health Research and Development, Ministry of Health, Republic of Indonesia | Viral vector (replicating) + APC | Phase 1/2 |
| COVI-VAC | Codagenix/Serum Institute of India | Live attenuated virus | Phase 1 |
| CIGB-669 (RBD+AgnHB) | Center for Genetic Engineering and Biotechnology | Protein subunit | Phase 1/2 |
| CIGB-66 (RBD + aluminium hydroxide) | Center for Genetic Engineering and Biotechnology | Protein subunit | Phase 1/2 |
| VLA2001 | Valneva + National Institute for Health Research, United Kingdom | Inactivated | Phase 1/2 |
| BECOV2 | Biological E., Ltd. | Protein subunit | Phase 1/2 |
| AdCLD-CoV19 | Cellid Co. Ltd. | Viral vector (replicating) | Phase 1/2 |
| GLS-5310 | GeneOne Life Science, Inc. | DNA | Phase 1/2 |
| Recombinant SARS-CoV-2 spike protein, aluminium adjuvanted | Nanogen Pharmaceutical Biotechnology | Protein subunit | Phase 1/2 |
| S-268019 | Shionogi Co., Ltd. | Protein subunit | Phase 1/2 |
| AdCOVID | Altimmune, Inc. | Viral vector | Phase 1 |
| SARS-CoV-2-RBD-Fc fusion protein | University Medical Center Groningen + Akston Biosciences Inc. | Protein subunit | Phase 1/2 |
| ERUCOV-VAC | Erciyes University | Inactivated | Phase 1 |
Note: This table is adapted from the list of vaccines currently undergoing clinical trials published by the World Health Organization,2 organized by candidate, company, mechanism, and phase of the clinical trial. This table is up to date as of January 6, 2021. Ad5: adenovirus type 5 vector; COVID-19: coronavirus disease 2019; LNP: lipid nanoparticle; RBD: receptor-binding domain; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
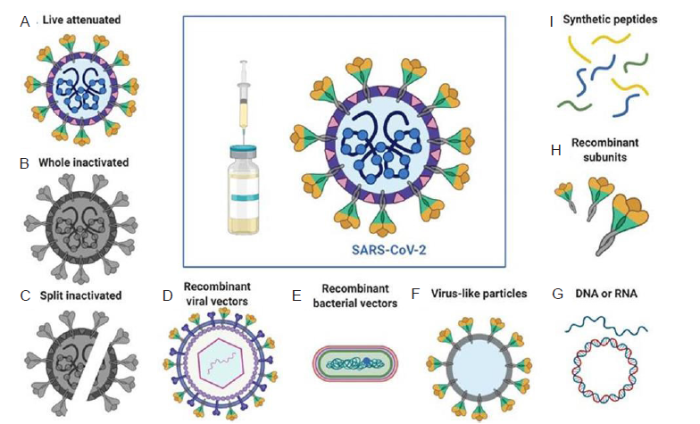
Inactivated virus and live attenuated virus vaccines are well-established means of conferring protection against a novel pathogen. Chemical, temperature, and radiation treatments are used to “inactivate” viruses by altering proteins involved in pathogenesis or preventing genome reading, while antigen epitopes remain intact to stimulate an immune response.11 Several inactivated virus vaccines are currently undergoing Phase 3 clinical trials or have emergency use authorization, primarily in China, as well as QazCovid-in in Kazakhstan.2 Live attenuated viruses, on the other hand, are created by propagating viruses under novel conditions that render them less pathogenic and less virulent. The added mutations which arise when growing under these conditions leads to an attenuated strain; however, there is still potential for the attenuated strain to revert back to the virulent strain, which makes them less safe than other vaccine technologies.12 Only one vaccine candidate currently in clinical trials uses the live attenuated virus, which is produced by Codagenix/Serum Institute of India.2 As vaccines of this type have been extensively studied, and many licensed inactivated or live attenuated virus vaccines exist, we will instead focus on vaccines made with novel biotechnologies, particularly, protein subunit, viral vector, mRNA, and DNA vaccines. Figure 3 summarizes the mechanisms of several of these vaccine technologies.13
Figure 3.
Figure 3.
Summary of SARS-CoV-2 vaccine types. A summary of several of the major vaccine types being manufactured, including live attenuated (A), inactivated (B, C), viral vector (D), bacterial vector (E), virus-like particles (F), DNA- or RNA-based (G), recombinant protein subunit (H), and synthetic peptides vaccines (I). Figure reprinted from Liu et al.13 Licensed under CC BY 4.0.
Recombinant Protein-based vaccines
As mentioned earlier, protein subunit vaccines are the most frequently-chosen vaccine type among the candidates currently undergoing clinical trials.2 Protein subunit vaccines, instead of using the whole virus, often utilize a specific antigenic protein. In the case of SARS-CoV-2, this is often a recombinant form of the full-length S protein, or specific domains on the S protein, such as the RBD.14
For instance, Novavax, which is testing a protein subunit-based SARS-CoV-2 vaccine in a Phase 3 study, utilizes a recombinant form of the full-length spike protein in conjunction with a Matrix-M1 adjuvant.15 The recombinant spike protein includes a mutation in the furin-cleavage site as well as two proline substitutions at residues K986P and V987P in order to prevent cleavage into the post-fusion form.15 This keeps epitopes present in the pre-fusion conformation accessible, allowing them to elicit neutralizing antibody responses.16 These mutations are made to the S-gene through cloning via the baculovirus expression system for expression in SF9 cells prior to extraction and purification.17 Anhui Zhifei Longcom Biopharmaceuticals, working with the Institute of Microbiology, Chinese Academy of Sciences, also has a protein subunit vaccine candidate currently under Phase 2 study.2 Their vaccine, however, uses a recombinant dimeric RBD.14
One hindrance in the development of protein subunit vaccines is that they display low immunogenicity without the addition of adjuvants.18 Thus, adjuvants need to be added to protein subunit vaccines in order to promote strong humoral and cellular immune responses. As mentioned earlier, the Novavax vaccine uses the Matrix-M1 adjuvant along with its recombinant spike protein subunit. Matrix-M1 is a saponin-based adjuvant that has been found to upregulate major histocompatibility complex class II as well as induce the recruitment and activation of dendritic cells, which go on to activate humoral and cellular immune responses.19, 20 A more extensive discussion of other adjuvants used in protein subunit vaccines can be found later in this paper.
Another biomaterial that has seen use in conjunction with protein subunits in SARS-CoV-2 vaccine candidates is virus-like particles (VLPs). VLPs are highly-structured arrangements of proteins from the viral capsid that mimic the virus structure but do not contain actual genetic material.14 VLPs have been shown to elicit both B cell and cytotoxic T cell immune responses, and as a result of mimicking viral structure, they often require lower doses of antigen than vaccines consisting of the protein subunit alone.21 In order to be safely mass produced, VLPs must be formed through an expression system, such as with hepatitis B virus in yeast cells or the baculovirus expression system that utilizes certain lepidopteran species.21 The nanoparticles that display the modified S protein subunit in the Novavax vaccine are one example of such VLPs.22 A vaccine produced by Medicago Inc. in Phase 2/3 clinical trials also uses a VLP produced in tobacco to display a recombinant SARS-CoV-2 spike protein.18
Viral vector vaccines
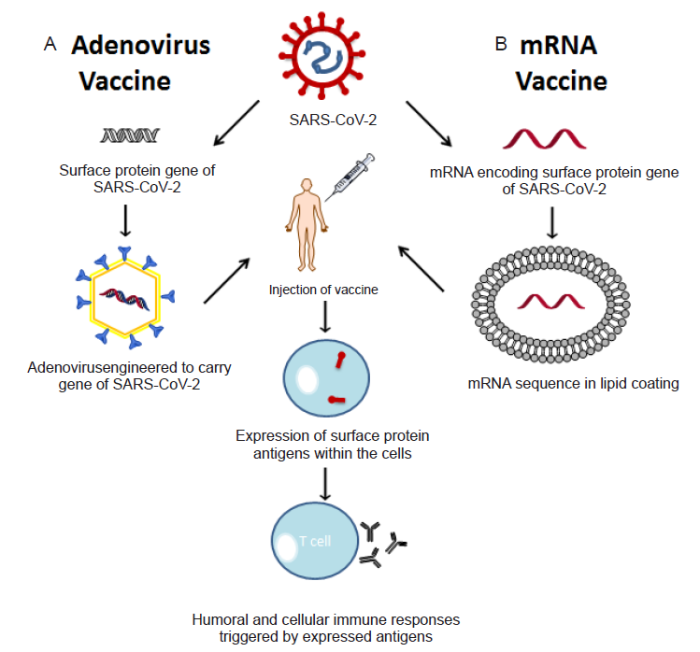
Viral vector-based vaccines are an emerging technology that clone specific SARS-CoV-2 antigens into the genetic material of either replicating or non-replicating virus vectors.23 Several non-replicating vector vaccines have entered Phase 3 clinical trials, and some have gained emergency use authorization in certain regions. These vaccines utilize adenovirus vectors with E1 gene deletions, preventing replication.22 Such vaccines currently under Phase 3 clinical trials are being produced by AstraZeneca/University of Oxford in the United Kingdom, Gamaleya Research Institute in Russia, CanSino Biologics, Inc. in China, and Janssen Pharmaceuticals, Inc. in the USA.2 Vaccines based on viral vectors work by transducing the antigenic gene via the vector into the host cell nucleus, where the gene is transcribed and later exported back into the cytoplasm to be translated and to elicit an immune response (Figure 4A).22 These vaccines hold potential to induce a highly-specific and efficient immune response against SARS-CoV-2.
Figure 4.
Figure 4.
Schematic mechanism of manufacturing of viral vector vaccines (A, adenovirus as example) and mRNA vaccines (B). The RNA of SARS-CoV-2 was sequenced, which identified the coding of surface proteins. Using endonuclease methods, an engineered mutated adenovirus vector that carries the SARS-CoV-2 surface protein gene was made. Different from the preparation of adenovirus, the mRNA sequences that encode the spike protein were directly generated. To enhance the stabilities of mRNA and to escape from human immunities, lipid nanoparticles were used to envelope the mRNA. After injection of both viral vector and mRNA vaccines, cells will read the mRNA sequence express the epitope of the surface protein (red within cell) in the cytoplasm or in the nucleus. This will trigger the host’s humoral and cellular immune responses that could potentially contribute to specific immunity to SARS-CoV-2.
Of the four viral vector-based vaccines currently in Phase 3 clinical trials, all use some form of a non-replicating adenovirus vector. Adenoviruses have double-stranded DNA and cause common cold symptoms in humans.22 Adenovirus vectors have several features that make them an attractive choice for vaccine developers. First, adenovirus vectors stimulate potent innate and adaptive immune responses while maintaining a high safety profile.24 Furthermore, transgenes can be inserted into adenovirus genomes, allowing for the expression of the target peptide as well as other immune response enhancers, such as cytokines and danger signals.24 The development of this biotechnology allows for high adaptability and can be exploited to increase the speed at which vaccines are produced. Also of interest is the separation of cellular attachment and entry processes in adenovirus vectors.24 The proteins responsible for recognition and attachment to certain receptors on host cells can be altered to increase specificity for receptors elsewhere without disrupting viral entry or gene transduction. Current SARS-CoV-2 vaccine candidates utilize both human and non-human adenovirus vectors.
For instance, CanSino Biologics, Inc. utilizes a recombinant human adenovirus type 5 vector (Ad5) in their vaccine, and AstraZeneca/University of Oxford’s AZD1222 vaccine uses the recombinant chimpanzee ChAdOx1 adenovirus vector. The Gamaleya-produced vaccine uses a combination of recombinant Ad5 and Ad26, while Janssen solely uses the Ad26 vector.14 All of these vaccines use the adenovirus vector to carry the full-length spike glycoprotein, where it is produced using the host cell’s machinery to be recognized and presented by antigen-presenting cells (APCs) to induce an immune response. The Janssen vaccine differs from the other three that use wild-type spike protein in that the S protein contains proline substitutions at K986P and V987P and two furin cleavage site mutations.23 One limitation to the adenovirus vector is the potential for immunity to certain vectors as a result of previous exposure. Indeed, a Phase 2 trial by CanSino Biologics, Inc. of their Ad5-vectored vaccine found that 52% of study participants had high pre-existing immunity to the Ad5 vector, which resulted in a two-fold decrease in neutralizing antibodies compared to those with minimal pre-existing immunity.25 A possible solution to this is using adenovirus vectors that have a lower seroprevalence in humans, such as Ad26 as used by Gamaleya Research Institute and Janssen Pharmaceuticals, Inc., or using a non-human adenovirus vector with very low human seroprevalence, as used by AstraZeneca/University of Oxford.26, 27
Although the majority of SARS-CoV-2 vaccine candidates using a viral vector mechanism in clinical trials are non-replicating and use adenovirus vectors, it is worth briefly discussing the candidates that use a replicating vector and/or non-adenovirus vector. A notable vaccine candidate of this type includes TMV-083, which is being produced by the Institut Pasteur in conjunction with Themis, Merck & Co. Inc., the University of Pittsburgh, and Merck Sharp & Dohme Ltd., and uses the measles virus as a vector.2 Recent studies have shown the measles vector platform, based on the established measles vaccine, to be safe and effective in Phase 1 and 2 clinical trials in formulating a vaccine against Chikungunya virus.28 Furthermore, current evidence shows that pre-existing immunity does not affect the vaccine functionality, indicating the potential of this viral vector to rapidly formulate a vaccine against novel pathogens.28
mRNA-based vaccines
Despite their novelty, several mRNA-based vaccine candidates have been developed and are currently undergoing clinical trials for SARS-CoV-2.2 mRNA-based vaccines offer high flexibility and adaptability, which allow them to be rapidly developed in the face of emerging pandemics.29 Indeed, the first two vaccines to receive emergency use authorizations from the United States Food and Drug Administration were mRNA-based vaccines produced by BioNTech/Pfizer and Moderna/NIAID. mRNA-based vaccines also offer the advantage of being self-adjuvanting. It has been shown that stabilized mRNA carries the ability to activate Toll-like receptors 7/8 and 3, which are essential for a primed immune response against viral targets.22 The innate immunostimulatory properties of mRNA can be utilized to elicit immune responses without the addition of an adjuvant, which can save resources by avoiding the need for additional safety testing or studying synergistic effects. Finally, mRNA vaccines only require the nucleic acid-encoded antigen to reach the cytosol of the target cells for translation to occur. This provides an additional safety element, especially compared to other nucleic acid-based vaccines, as there is no potential for integration into the genome (Figure 4B).18
Two limitations to RNA-based vaccines are the inherent instability of mRNA in vivo and the low translatability of “naked” mRNA. Several strategies have been developed in order to circumvent this issue and deliver the antigenic RNA without rapid degradation by RNases. Stabilization of mRNA can be achieved through modifications to the 5′- and 3′-untranslated region elements, which surround the ORF containing the antigenic gene.30 These modifications include synthetically adding a 5′ cap, regulating the poly(A) tail length, and optimizing codon sequences.29 In addition to stabilizing mRNA, these modifications can increase protein translation.30 Another modification of interest is the use of protamine, a polycationic peptide that protects mRNA from degradation.31 Protamine, however, shows limited efficacy when complexing mRNA in and of itself, but efficacy is improved when it is included as part of an mRNA vaccine platform.30 mRNA vaccine platforms often include encapsulation by lipid nanoparticles (LNPs). LNPs contain ionizable cationic lipids that aid in vivo delivery of mRNA to target cells.32
Due to the high adaptability of mRNA-based vaccines, various approaches have been taken to develop such a vaccine against SARS-CoV-2. For instance, the mRNA-1273 vaccine produced by Moderna/NIAID encodes the full-length, pre-cleavage stabilized spike protein within an LNP capsule.18 Four lipids are used in a fixed ratio with the mRNA, although the exact composition of the lipids is unknown.33 BNT162, the mRNA vaccine candidate produced by BioNTech/Pfizer, uses nucleoside-modified RNA that encodes the RBD of the SARS-CoV-2 spike protein.34 The addition of 1-methyl-pseudouiridine has been found to reduce the immunogenicity of mRNA, while increasing stability and protein translation.35 Additionally, BNT162 utilizes a T4 fibritin-derived “foldon” trimerization domain, which allows for a multivalent display of the RBD antigen, thus increasing the number of binding sites and immunogenicity.34 Like the Moderna/NIAID vaccine, BNT162 is encapsulated within LNPs and does not mention any use of adjuvant. Both vaccine formulations have been found to cause minimal negative side effects and high efficacy thus far.36, 37
DNA vaccines
Like RNA-based vaccines, DNA vaccines utilize genetic material that codes for specific antigenic proteins on SARS-CoV-2 and can be rapidly developed against novel pathogens for mass production. Likewise, DNA-based vaccines work in a similar manner to mRNA-based vaccines. The antigen is encoded by a sequence incorporated into a DNA plasmid, which is then transfected into host cells. There, host machinery is used to transcribe and translate the antigen into a functional peptide.23 The use of DNA rather than mRNA comes with both advantages and disadvantages. For instance, while mRNA is intrinsically unstable and can be degraded by RNases, DNA offers greater stability meaning DNA expression is longer-lived, thus potentially conferring a more potent immune response, and cold chain transport is not required.18 However, a major disadvantage to the use of DNA-based vaccines is the potential for host genome integration, as the antigenic DNA must enter the host cell nucleus to be transcribed.18
Although several DNA-based vaccines are currently undergoing clinical trials, to the best of the authors’ knowledge only Inovio Pharmaceuticals has begun Phase 3 clinical trials and published data on their INO-4800 vaccine.38 Inovio Pharmaceuticals, which is also currently testing a DNA-based vaccine against MERS-CoV, developed their SARS-CoV-2 vaccine INO-4800 to encode the full-length spike glycoprotein along with an N-terminal IgE leader sequence. This optimized DNA sequence is encoded on a plasmid labelled pGX9501 and has been shown to elicit both cellular and humoral responses against the spike protein following immunization of mice and guinea pigs.38 One interesting aspect of INO-4800 is the use of electroporation to administer the vaccine intradermally. Electroporation is an interesting biotechnology that uses short electrical pulses to increase cell membrane permeability and pDNA uptake at the vaccine administration site, which has been associated with a greater recruitment of APCs and inflammatory cells.39
Another interesting DNA-based vaccine, bacTRL-Spike, has been developed by Symvivo Corporation and is currently undergoing a Phase 1 clinical trial set to be completed in February 2022 (NCT04334980). The bacTRL-spike vaccine, which is taken orally, marks the first in-human use of the Bifidobacterium longum vector to deliver a modified DNA plasmid containing the SARS-CoV-2 spike protein. B. longum is an anaerobic bacterium present in the human microbiome; therefore, it does not present a risk for virulence.39 Additionally, strains of B. longum have previously been tested as carriers of hepatitis C virus and enterovirus, but not in human hosts.39
Importance of Formulation
Adjuvants
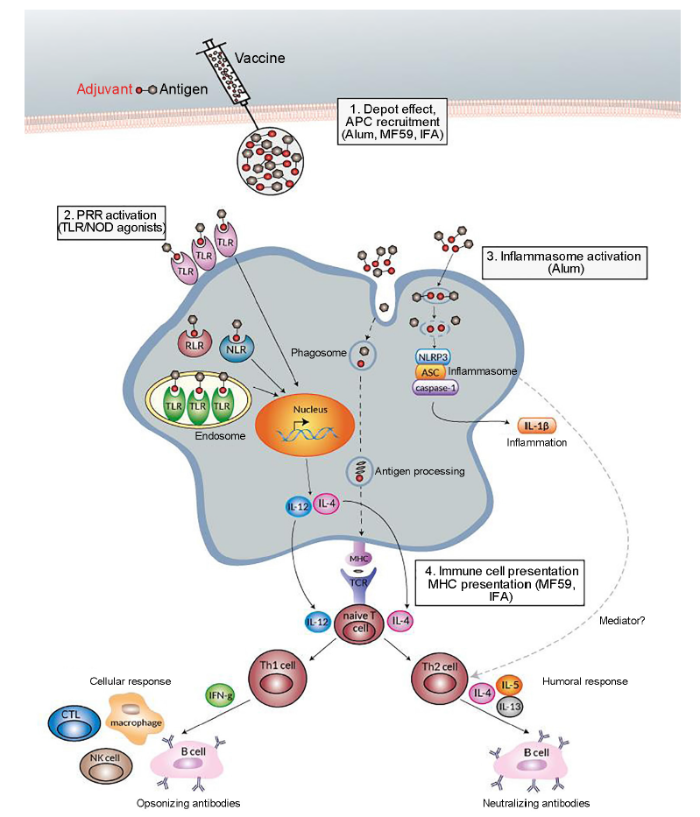
As mentioned earlier, adjuvants are immunostimulatory agents that are often added to vaccines to improve the ability of antigens to induce an immune response. While nucleic acid-based vaccines are considered self-adjuvanting given their high immunogenicity, and viral vectors prime the immune response through the vector, protein subunit vaccines require the use of adjuvants.22 Some adjuvants that are seeing use in the development of protein subunit-based SARS-CoV-2 vaccines include alum, Matrix-M1, and CpG.2 Figure 5 summarizes the various mechanisms of adjuvants for improving high immunogenicity.
Figure 5.
Figure 5.
Adjuvants improve immunogenicity via different mechanisms. 1. Alum and emulsion such as MF59 generate depots to trap and recruit antigen presenting cells (APCs). 2. By utilizing TLR/NOD agonists, pattern recognition receptors (PRR) were covalently bound to their ligands, followed by the activation of downstream pathways. 3. Aside from APC recruitment, Alum could also induce NLRP3 inflammasome. 4. Depot generation and induction of MHC responses could be obtained by application of MF59 and Freund’s Incomplete Adjuvant (IFA). The image is licensed and authorized by InvivoGen.
Alum is an aluminium-based adjuvant that has a long history of use as a clinical adjuvant. The addition of alum adjuvants promotes the adaptive immune response through uric acid, which induces the differentiation of dendritic cells.40 Several current vaccines against COVID-19 utilize alum adjuvants, such as SCB-2019 and CIGB-66.2 Matrix-M1, which was discussed in the context of the Novavax protein subunit vaccine, is a saponin-based adjuvant in an immune-stimulating complex-matrix conformation. This conformation includes a specific fraction of saponin, cholesterol, phospholipids, and the antigen of choice, with the Matrix-M1 adjuvant including a mix of two different matrices (Matrix-A and Matrix-C) that have different saponin fractions.19 Among the vaccines currently undergoing clinical trials, only Novavax uses the Matrix-M1 adjuvant to the best of our knowledge.2 An interesting adjuvant is the use of CpG, which consists of unmethylated CG dinucleotides derived from bacterial DNA.41 As CpG is expressed more highly in bacteria than eukaryotes, it is naturally recognized by Toll-like receptor-9 to trigger an innate immune response.41 As DNA-based vaccines use recombinant bacterial DNA, they naturally contain CpG sequences which promote the innate immune response. Protein subunit-based vaccines currently in clinical trials, like SCB-2019 and MVC-COV1901, also exploit the use of these sequences to boost their immunogenicity.2
Cold chain transport
Since the widespread use of vaccination as a public health measure in the 1960s and 1970s, the necessity for a “vaccine cold chain” to transport temperature-sensitive vaccines has been underscored. Difficulties in storing and shipping these vaccines is particularly the case in tropical climates, where electricity is unstable, appropriate equipment is unavailable, and there is a lack of sufficiently trained staff.42 However, these issues are primarily true for inactivated and live attenuated vaccines, which require storage at approximately 2°C to 8°C, with the exception of varicella vaccines which require storage at -50°C to -15°C.43 In contrast, many of the new mRNA-based vaccines developed during the COVID-19 pandemic require storage at temperatures below these ranges. For instance, the BioNTech/Pfizer mRNA-based vaccine requires storage and transport at temperatures as low as -80°C to -60°C to remain stable for up to 6 months, while the Moderna/NIAID vaccine requires storage at -20°C for up to 6 months.44 While stability at the more attractive 2°C to 8°C is possible for brief periods of time (5 days for Pfizer/BioNTech and 30 days for Moderna/NIAID), this requires that all the vaccine doses are used quickly and presents a problem in developing countries where such freezers are not available.44
As mentioned by Crommelin et al.44 in their review of mRNA vaccine thermostability, liquid and lyophilized formulations of mRNA vaccines could provide refrigerated stability. One mRNA vaccine candidate, referred to as ARCov, uses a liquid formulation to deliver the LNP-encapsulated antigenic mRNA. A study using mice demonstrated that this vaccine induces both neutralizing antibodies and T-cell immune responses, as well as displaying thermostability at 2°C and 25°C for up to a week, though the authors acknowledge that the persistence of neutralizing antibodies is known as well as long-term thermostability at 2°C and 25°C.45 Participants are currently being recruited for a Phase 1 study of this vaccine at Shulan Hospital.2 Other vaccine formulations, such as those with viral vectors or DNA, also provide the high adaptability and scalability of mRNA-based vaccines combined with greater thermostability, which may aid in their distribution in developing countries to fight the global COVID-19 pandemic.15, 46
Summary and Perspective
The global COVID-19 pandemic has seen the adoption of several novel technologies in vaccine development as companies race to produce and deliver a safe and effective vaccine against SARS-CoV-2. Although conventional inactivated and live attenuated vaccines are being produced and approved by many countries, the COVID-19 pandemic has notably provided the opportunity to utilize protein subunit, viral vector, mRNA-, and DNA-based vaccine technologies due to their high adaptability and potential to be scaled up rapidly. Additionally, the development of nanoscale biomaterials has greatly enhanced the delivery, immunogenicity, and safety of these novel vaccines. As discussed earlier, the design of VLPs to mimic live or inactivated viruses has helped to increase the potency of immune responses for protein subunit vaccines, as well as nanoparticle-based adjuvants like Matrix-M1.21 LNPs, used to encapsulate the mRNA antigen, aid in the delivery to target cells as well as the stability of mRNA, thereby increasing the potency of these vaccines through reduced mRNA degradation and increased protein translation. Modifications to the mRNA nucleotide sequence, particularly in the untranslated regions, can also improve stability and decrease innate immunogenicity that could trigger inflammation and other severe immune responses.47
Despite the many successes in the development of biomaterials, several avenues of research remain to be utilized in the rapid formulation, testing, production, and distribution of vaccines against novel pathogenic agents. First, despite their scalability and potential to be rapidly developed, the distribution of mRNA-LNP vaccines in developing nations is hindered by their instability and the requirement for the “cold chain” for vaccine distribution. Further research and development would be warranted in improving mRNA vaccine thermostability while retaining safety and efficacy, such as through lyophilized or liquid formulations.44 Additionally, just as the development of refrigerators and thermal sensors to monitor the status of vaccines was necessary for the eradication of smallpox and the ongoing effort to eradicate polio, the formulation of ultra-cold freezers that can be adapted to developing nations is necessary for future widespread adoption of mRNA-based vaccines.42
Secondly, much potential remains in the application of nanobiotechnology to increase the structure and polyvalency of vaccine platforms. The seminal study by Bachmann et al.48 demonstrated that high-density, organized antigen displays resulted in higher IgM titres and created better B cell activation in transgenic mice compared to less-ordered displays. These results highlight the potential for highly-ordered scaffolds, such as virus nanoparticles and VLPs, for application in presenting organized antigens that mimic the pathogen. An interesting recent study showed that rod-like viral particles outperformed icosahedral viral scaffolds in eliciting a long-lasting immune response when small and weakly-immunogenic haptens were displayed on the external surface of the viral capsids.49 Additionally, it is possible to apply self-assembling polymeric particles to present high-density antigens to enhance the immune responses.50 Equally relevant is the importance of polyvalent interactions between the selected antigen and APCs during antigen recruitment. Such interactions have been found to be stronger than their monovalent counterparts, increase the biological lifetime of the polyvalent molecules, and aid in the binding specificity of receptors to particular ligands.51 Therefore, using bioconjugation technologies, a highly structured, polyvalent antigen presentation can be designed on the surfaces of VLPs or similar polyvalent scaffolds to boost immunogenicity and improve immune response of proteins or small molecular antigens.
Thirdly, as the COVID-19 pandemic marks the first time mRNA, DNA, and viral vector vaccines are seeing widespread use in humans, there is some hesitation on the safety of these vaccines. According to a study by the Pew Research Center in September 2020, only 51% of U.S. adults who responded stated that they would get the vaccine if it were available, and only 21% responded that they would definitely get vaccinated.52 Although a variety of factors affect the reception of vaccines, including political and religious beliefs, demonstrating long-term safety and efficacy is essential for widespread adoption. With the rigorous and extensive clinical testing these novel vaccines are receiving around the world, their long-term safety will likely be demonstrated in the years to come. Nonetheless, it will be useful to develop methodologies that can more rapidly determine long-term efficacy and safety.
Furthermore, the recent announcement of the emergence of SARS-CoV-2 strains with increased infectivity in South Africa and the United Kingdom have heralded some worries over the efficacy of the newly-developed vaccines.53, 54 While both have mutations in the spike protein, which is the target of many vaccines in development and clinical trials, it is likely the vaccines will still work, as they bind to multiple epitopes to induce protection. In the case that key epitopes contain the mutations, the adaptability of these vaccines should allow them to be quickly modified to provide protection against these strains as well.
Finally, a plethora of bionanotechnologies has been utilized to produce safe and efficacious vaccines against SARS-CoV-2. Advances in these technologies allowed for their development and deployment against a novel pathogen at record speed. Vaccine platforms, such as LNP-encapsulated nucleic acid sequences, non-pathogenic viral vectors, and protein subunits, have a high degree of scalability and adaptability that will allow them to be readily put to use against future strains of SARS-CoV-2 or other novel pathogens. Biomaterials research should seek to utilize innovative technologies to enhance the immunogenicity and stability of vaccines while reducing deleterious reactions. Future biomaterials research should focus on developing novel adjuvants that improve safety profiles while heightening immune response, improving efficient interaction of nanoparticles with APCs, and generating expression systems that improve scalability and distribution in developing nations.55
In summary, the future seems bright for the development and application of novel vaccination strategies. Nonetheless, the continued refinement and development of nanotechnologies and biomaterials to modify these vaccines is warranted in order to improve their safety, efficacy, immunogenicity, and delivery to combat emerging strains of SARS-CoV-2 and prevent future pandemics.
Author contributions
IJ and JF provided the concept and design of the review. IJ was responsible for searching the literature and manuscript preparation. Both IJ and JF participated in manuscript editing and manuscript review. Both authors approved the final version of this manuscript.
Financial support
The authors received no funding for this review.
Acknowledgement
None.
Conflicts of interest statement
The authors declare no competing financial interests.
Data sharing statement
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Reference
Characteristics of SARS-CoV-2 and COVID-19
DOI:10.1038/s41579-020-00459-7
URL
PMID:33024307
[Cited within: 5]

Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a highly transmissible and pathogenic coronavirus that emerged in late 2019 and has caused a pandemic of acute respiratory disease, named 'coronavirus disease 2019' (COVID-19), which threatens human health and public safety. In this Review, we describe the basic virology of SARS-CoV-2, including genomic characteristics and receptor use, highlighting its key difference from previously known coronaviruses. We summarize current knowledge of clinical, epidemiological and pathological features of COVID-19, as well as recent progress in animal models and antiviral treatment approaches for SARS-CoV-2 infection. We also discuss the potential wildlife hosts and zoonotic origin of this emerging virus in detail.
Severe Acute Respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response
DOI:10.1016/j.dsx.2020.04.020
URL
PMID:32335367
[Cited within: 1]

BACKGROUND AND AIM: As a result of its rapid spread in various countries around the world, on March 11, 2020, WHO issued an announcement of the change in coronavirus disease 2019 status from epidemic to pandemic disease. The virus that causes this disease is indicated originating from animals traded in a live animal market in Wuhan, China. Severe Acute Respiratory Syndrome Coronavirus 2 can attack lung cells because there are many conserved receptor entries, namely Angiotensin Converting Enzyme-2. The presence of this virus in host cells will initiate various protective responses leading to pneumonia and Acute Respiratory Distress Syndrome. This review aimed to provide an overview related to this virus and examine the body's responses and possible therapies. METHOD: We searched PubMed databases for Severe Acute Respiratory Syndrome Coronavirus-2, Middle East respiratory syndrome-related coronavirus and Severe Acute Respiratory Syndrome Coronavirus. Full texts were retrieved, analyzed and developed into an easy-to-understand review. RESULTS: We provide a complete review related to structure, origin, and how the body responds to this virus infection and explain the possibility of an immune system over-reaction or cytokine storm. We also include an explanation of how this virus creates modes of avoidance to evade immune system attacks. We further explain the therapeutic approaches that can be taken in the treatment and prevention of this viral infection. CONCLUSION: In summary, based on the structural and immune-evasion system of coronavirus, we suggest several approaches to treat the disease.
COVID-19 pandemic crisis-a complete outline of SARS-CoV-2
DOI:10.1186/s43094-020-00133-y
URL
PMID:33224993
[Cited within: 2]

Background: Coronavirus (SARS-CoV-2), the cause of COVID-19, a fatal disease emerged from Wuhan, a large city in the Chinese province of Hubei in December 2019. Main body of abstract: The World Health Organization declared COVID-19 as a pandemic due to its spread to other countries inside and outside Asia. Initial confirmation of the pandemic shows patient exposure to the Huanan seafood market. Bats might be a significant host for the spread of coronaviruses via an unknown intermediate host. The human-to-human transfer has become a significant concern due to one of the significant reasons that is asymptomatic carriers or silent spreaders. No data is obtained regarding prophylactic treatment for COVID-19, although many clinical trials are underway. Conclusion: The most effective weapon is prevention and precaution to avoid the spread of the pandemic. In this current review, we outline pathogenesis, diagnosis, treatment, ongoing clinical trials, prevention, and precautions. We have also highlighted the impact of pandemic worldwide and challenges that can help to overcome the fatal disease in the future.
Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19
DOI:10.1038/s41401-020-0485-4
URL
PMID:32747721
[Cited within: 3]

Coronavirus disease 2019 is a newly emerging infectious disease currently spreading across the world. It is caused by a novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The spike (S) protein of SARS-CoV-2, which plays a key role in the receptor recognition and cell membrane fusion process, is composed of two subunits, S1 and S2. The S1 subunit contains a receptor-binding domain that recognizes and binds to the host receptor angiotensin-converting enzyme 2, while the S2 subunit mediates viral cell membrane fusion by forming a six-helical bundle via the two-heptad repeat domain. In this review, we highlight recent research advance in the structure, function and development of antivirus drugs targeting the S protein.
SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals
DOI:10.1038/s41598-020-71936-5
URL
PMID:33020502
[Cited within: 2]

SARS-CoV-2 has a zoonotic origin and was transmitted to humans via an undetermined intermediate host, leading to infections in humans and other mammals. To enter host cells, the viral spike protein (S-protein) binds to its receptor, ACE2, and is then processed by TMPRSS2. Whilst receptor binding contributes to the viral host range, S-protein:ACE2 complexes from other animals have not been investigated widely. To predict infection risks, we modelled S-protein:ACE2 complexes from 215 vertebrate species, calculated changes in the energy of the complex caused by mutations in each species, relative to human ACE2, and correlated these changes with COVID-19 infection data. We also analysed structural interactions to better understand the key residues contributing to affinity. We predict that mutations are more detrimental in ACE2 than TMPRSS2. Finally, we demonstrate phylogenetically that human SARS-CoV-2 strains have been isolated in animals. Our results suggest that SARS-CoV-2 can infect a broad range of mammals, but few fish, birds or reptiles. Susceptible animals could serve as reservoirs of the virus, necessitating careful ongoing animal management and surveillance.
Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target
DOI:10.1007/s00134-020-05985-9 URL PMID:32125455 [Cited within: 1]
Correlates of protection against SARS-CoV-2 in rhesus macaques
DOI:10.1038/s41586-020-03041-6
URL
PMID:33276369
[Cited within: 2]

Recent studies have reported the protective efficacy of both natural(1) and vaccine-induced(2-7) immunity against challenge with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in rhesus macaques. However, the importance of humoral and cellular immunity for protection against infection with SARS-CoV-2 remains to be determined. Here we show that the adoptive transfer of purified IgG from convalescent rhesus macaques (Macaca mulatta) protects naive recipient macaques against challenge with SARS-CoV-2 in a dose-dependent fashion. Depletion of CD8(+) T cells in convalescent macaques partially abrogated the protective efficacy of natural immunity against rechallenge with SARS-CoV-2, which suggests a role for cellular immunity in the context of waning or subprotective antibody titres. These data demonstrate that relatively low antibody titres are sufficient for protection against SARS-CoV-2 in rhesus macaques, and that cellular immune responses may contribute to protection if antibody responses are suboptimal. We also show that higher antibody titres are required for treatment of SARS-CoV-2 infection in macaques. These findings have implications for the development of SARS-CoV-2 vaccines and immune-based therapeutic agents.
Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study
DOI:10.4049/jimmunol.0903490
URL
PMID:21576510
[Cited within: 1]

Six years have passed since the outbreak of severe acute respiratory syndrome (SARS). Previous studies indicated that specific Abs to SARS-related coronavirus (SARS-CoV) waned over time in recovered SARS patients. It is critical to find out whether a potential anamnestic response, as seen with other viral infections, exists to protect a person from reinfection in case of another SARS outbreak. Recovered SARS patients were followed up to 6 y to estimate the longevity of specific Ab. The specific memory B cell and T cell responses to SARS-CoV Ags were measured by means of ELISPOT assay. Factors in relation to humoral and cellular immunity were investigated. Six years postinfection, specific IgG Ab to SARS-CoV became undetectable in 21 of the 23 former patients. No SARS-CoV Ag-specific memory B cell response was detected in either 23 former SARS patients or 22 close contacts of SARS patients. Memory T cell responses to a pool of SARS-CoV S peptides were identified in 14 of 23 (60.9%) recovered SARS patients, whereas there was no such specific response in either close contacts or healthy controls. Patients with more severe clinical manifestations seemed to present a higher level of Ag-specific memory T cell response. SARS-specific IgG Ab may eventually vanish and peripheral memory B cell responses are undetectable in recovered SARS patients. In contrast, specific T cell anamnestic responses can be maintained for at least 6 y. These findings have applications in preparation for the possible reemergence of SARS.
Antibodies, immunity, and COVID-19
DOI:10.1001/jamainternmed.2021.0074 URL PMID:33720309 [Cited within: 1]
Inactivated virus vaccines from chemistry to prophylaxis: merits, risks and challenges
DOI:10.1586/erv.12.38
URL
PMID:22873127
[Cited within: 1]

The aim of this review is to make researchers aware of the benefits of an efficient quality control system for prediction of a developed vaccine's efficacy. Two major goals should be addressed when inactivating a virus for vaccine purposes: first, the infectious virus should be inactivated completely in order to be safe, and second, the viral epitopes important for the induction of protective immunity should be conserved after inactivation in order to have an antigen of high quality. Therefore, some problems associated with the virus inactivation process, such as virus aggregate formation, protein crosslinking, protein denaturation and degradation should be addressed before testing an inactivated vaccine in vivo.
Evolutionary dynamics of viral attenuation
DOI:10.1128/jvi.76.20.10524-10529.2002
URL
PMID:12239331
[Cited within: 1]

The genetic trajectory leading to viral attenuation was studied in a canine parvovirus (CPV) strain grown on dog kidney cells for 115 transfers. Consensus sequences of viral populations at passages 0, 3, 30, 50, 80, and 115 were obtained from PCR products covering 86% of the genome; clones from each of the 80th and 115th passages were also sequenced, covering 69% of the genome. Sixteen changes were fixed in the 115th-passage virus sample. Levels of polymorphism were strikingly different over time, in part because of a plaque-cloning step at passage 112 that reduced variation: passage 80 had 19 variants common among the clones, but passage 115 had only a single common variant. Several mutations increased in the culture at the same time, with most reaching fixation only after the 80th passage. The pattern of evolution was consistent with recombination and not with separate selective sweeps of individual mutations. Thirteen of the changes observed were identical to or at the same positions as changes observed in other isolates of CPV or feline panleukopenia virus.
COVID-19: Progress in diagnostics, therapy and vaccination
DOI:10.7150/thno.47987
URL
PMID:32685022
[Cited within: 2]

Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has recently become a pandemic. As the sudden emergence and rapid spread of SARS-CoV-2 is endangering global health and the economy, the development of strategies to contain the virus's spread are urgently needed. At present, various diagnostic kits to test for SARS-CoV-2 are available for use to initiate appropriate treatment faster and to limit further spread of the virus. Several drugs have demonstrated in vitro activity against SARS-CoV-2 or potential clinical benefits. In addition, institutions and companies worldwide are working tirelessly to develop treatments and vaccines against COVID-19. However, no drug or vaccine has yet been specifically approved for COVID-19. Given the urgency of the outbreak, we focus here on recent advances in the diagnostics, treatment, and vaccine development for SARS-CoV-2 infection, helping to guide strategies to address the current COVID-19 pandemic.
A systematic review of SARS-CoV-2 vaccine candidates
DOI:10.1038/s41392-020-00352-y
URL
PMID:33051445
[Cited within: 4]

Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an emerging virus that is highly pathogenic and has caused the recent worldwide pandemic officially named coronavirus disease (COVID-19). Currently, considerable efforts have been put into developing effective and safe drugs and vaccines against SARS-CoV-2. Vaccines, such as inactivated vaccines, nucleic acid-based vaccines, and vector vaccines, have already entered clinical trials. In this review, we provide an overview of the experimental and clinical data obtained from recent SARS-CoV-2 vaccines trials, and highlight certain potential safety issues that require consideration when developing vaccines. Furthermore, we summarize several strategies utilized in the development of vaccines against other infectious viruses, such as severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), with the aim of aiding in the design of effective therapeutic approaches against SARS-CoV-2.
Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine
DOI:10.1056/NEJMoa2026920
URL
PMID:32877576
[Cited within: 3]

Rapid COVID-19 vaccine development
DOI:10.1126/science.abb8923 URL PMID:32385100 [Cited within: 1]
SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice
DOI:10.1038/s41467-020-20653-8
URL
PMID:33446655
[Cited within: 1]

The COVID-19 pandemic continues to spread throughout the world with an urgent need for a safe and protective vaccine to effectuate herd protection and control the spread of SARS-CoV-2. Here, we report the development of a SARS-CoV-2 subunit vaccine (NVX-CoV2373) from the full-length spike (S) protein that is stable in the prefusion conformation. NVX-CoV2373 S form 27.2-nm nanoparticles that are thermostable and bind with high affinity to the human angiotensin-converting enzyme 2 (hACE2) receptor. In mice, low-dose NVX-CoV2373 with saponin-based Matrix-M adjuvant elicit high titer anti-S IgG that blocks hACE2 receptor binding, neutralize virus, and protects against SARS-CoV-2 challenge with no evidence of vaccine-associated enhanced respiratory disease. NVX-CoV2373 also elicits multifunctional CD4(+) and CD8(+) T cells, CD4(+) follicular helper T cells (Tfh), and antigen-specific germinal center (GC) B cells in the spleen. In baboons, low-dose levels of NVX-CoV2373 with Matrix-M was also highly immunogenic and elicited high titer anti-S antibodies and functional antibodies that block S-protein binding to hACE2 and neutralize virus infection and antigen-specific T cells. These results support the ongoing phase 1/2 clinical evaluation of the safety and immunogenicity of NVX-CoV2373 with Matrix-M (NCT04368988).
COVID-19 vaccine: a comprehensive status report
DOI:10.1016/j.virusres.2020.198114
URL
PMID:32800805
[Cited within: 6]

The current COVID-19 pandemic has urged the scientific community internationally to find answers in terms of therapeutics and vaccines to control SARS-CoV-2. Published investigations mostly on SARS-CoV and to some extent on MERS has taught lessons on vaccination strategies to this novel coronavirus. This is attributed to the fact that SARS-CoV-2 uses the same receptor as SARS-CoV on the host cell i.e. human Angiotensin Converting Enzyme 2 (hACE2) and is approximately 79% similar genetically to SARS-CoV. Though the efforts on COVID-19 vaccines started very early, initially in China, as soon as the outbreak of novel coronavirus erupted and then world-over as the disease was declared a pandemic by WHO. But we will not be having an effective COVID-19 vaccine before September, 2020 as per very optimistic estimates. This is because a successful COVID-19 vaccine will require a cautious validation of efficacy and adverse reactivity as the target vaccinee population include high-risk individuals over the age of 60, particularly those with chronic co-morbid conditions, frontline healthcare workers and those involved in essentials industries. Various platforms for vaccine development are available namely: virus vectored vaccines, protein subunit vaccines, genetic vaccines, and monoclonal antibodies for passive immunization which are under evaluations for SARS-CoV-2, with each having discrete benefits and hindrances. The COVID-19 pandemic which probably is the most devastating one in the last 100 years after Spanish flu mandates the speedy evaluation of the multiple approaches for competence to elicit protective immunity and safety to curtail unwanted immune-potentiation which plays an important role in the pathogenesis of this virus. This review is aimed at providing an overview of the efforts dedicated to an effective vaccine for this novel coronavirus which has crippled the world in terms of economy, human health and life.
Matrix-MTM adjuvant induces local recruitment, activation and maturation of central immune cells in absence of antigen
DOI:10.1371/journal.pone.0041451
URL
PMID:22844480
[Cited within: 2]

Saponin-based adjuvants are widely used to enhance humoral and cellular immune responses towards vaccine antigens, although it is not yet completely known how they mediate their stimulatory effects. The aim of this study was to elucidate the mechanism of action of adjuvant Matrix-M without antigen and Alum was used as reference adjuvant. Adjuvant Matrix-M is comprised of 40 nm nanoparticles composed of Quillaja saponins, cholesterol and phospholipid. BALB/c mice were subcutaneously injected once with, 3, 12 or 30 microg of Matrix-M, resulting in recruitment of leukocytes to draining lymph nodes (dLNs) and spleen 48 h post treatment. Flow cytometry analysis identified CD11b(+) Gr-1(high) granulocytes as the cell population increasing most in dLNs and spleen. Additionally, dendritic cells, F4/80(int) cells, T-, B- and NK-cells were recruited to dLNs and in spleen the number of F4/80(int) cells, and to some extent, B cells and dendritic cells, increased. Elevated levels of early activation marker CD69 were detected on T-, B- and NK-cells, CD11b(+) Gr-1(high) cells, F4/80(int) cells and dendritic cells in dLNs. In spleen CD69 was mainly up-regulated on NK cells. B cells and dendritic cells in dLNs and spleen showed an increased expression of the co-stimulatory molecule CD86 and dendritic cells in dLNs expressed elevated levels of MHC class II. The high-dose (30 microg) of Matrix-M induced detectable serum levels of IL-6 and MIP-1beta 4 h post administration, most likely representing spillover of locally produced cytokines. A lesser increase of IL-6 in serum after administration of 12 microg Matrix-M was also observed. In conclusion, early immunostimulatory properties were demonstrated by Matrix-M alone, as therapeutic doses resulted in a local transient immune response with recruitment and activation of central immune cells to dLNs. These effects may play a role in enhancing uptake and presentation of vaccine antigens to elicit a competent immune response.
2 - Vaccine Immunology
In Plotkin’s Vaccines (Seventh Edition), Plotkin, S. A.; Orenstein, W. A.; Offit, P. A.; Edwards, K. M., eds.
Virus-like particles as immunogens
DOI:10.1016/s0966-842x(03)00208-7
URL
PMID:13678860
[Cited within: 3]

Subunit vaccines based on recombinant proteins can suffer from poor immunogenicity owing to incorrect folding of the target protein or poor presentation to the immune system. Virus-like particles (VLPs) represent a specific class of subunit vaccine that mimic the structure of authentic virus particles. They are recognized readily by the immune system and present viral antigens in a more authentic conformation than other subunit vaccines. VLPs have therefore shown dramatic effectiveness as candidate vaccines. Here, we review the current status of VLPs as vaccines, and discuss the characteristics and problems associated with producing VLPs for different viruses.
COVID-19 vaccine frontrunners and their nanotechnology design
Viral targets for vaccines against COVID-19
DOI:10.1038/s41577-020-00480-0
URL
PMID:33340022
[Cited within: 3]

Vaccines are urgently needed to control the coronavirus disease 2019 (COVID-19) pandemic and to help the return to pre-pandemic normalcy. A great many vaccine candidates are being developed, several of which have completed late-stage clinical trials and are reporting positive results. In this Progress article, we discuss which viral elements are used in COVID-19 vaccine candidates, why they might act as good targets for the immune system and the implications for protective immunity.
Development of an adenovirus vector vaccine platform for targeting dendritic cells
DOI:10.1038/s41417-017-0002-1
URL
PMID:29242639
[Cited within: 3]

Adenoviral (Ad) vector vaccines represent one of the most promising modern vaccine platforms, and Ad vector vaccines are currently being investigated in human clinical trials for infectious disease and cancer. Our studies have shown that specific targeting of adenovirus to dendritic cells dramatically enhanced vaccine efficacy. However, this was achieved using a molecular adapter, thereby necessitating a two component vector approach. To address the mandates of clinical translation of our strategy, we here sought to accomplish the goal of DC targeting with a single-component adenovirus vector approach. To redirect the specificity of Ad vector vaccines, we replaced the Ad fiber knob with fiber-fibritin chimeras fused to DC1.8, a single-domain antibody (sdAb) specific for murine immature DC. We engineered a fiber-fibritin-sdAb chimeric molecule using the coding sequence for DC1.8, and then replaced the native Ad5 fiber knob sequence by homologous recombination. The resulting Ad5 virus, Ad5FF1.8, expresses the chimeric fiber-fibritin sdAb chimera. Infection with Ad5FF1.8 dramatically enhances transgene expression in DC2.4 dendritic cells compared with infection with native Ad5. Ad5FF1.8 infection of bone marrow-derived DC demonstrates that Ad5FF1.8 selectively infects immature DC consistent with the known specificity of DC1.8. Thus, sdAb can be used to selectively redirect the tropism of Ad5 vector vaccines, providing the opportunity to engineer Ad vector vaccines that are specifically targeted to DC, or specific DC subsets.
Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial
DOI:10.1016/S0140-6736(20)31605-6
URL
PMID:32702299
[Cited within: 1]

BACKGROUND: This is the first randomised controlled trial for assessment of the immunogenicity and safety of a candidate non-replicating adenovirus type-5 (Ad5)-vectored COVID-19 vaccine, aiming to determine an appropriate dose of the candidate vaccine for an efficacy study. METHODS: This randomised, double-blind, placebo-controlled, phase 2 trial of the Ad5-vectored COVID-19 vaccine was done in a single centre in Wuhan, China. Healthy adults aged 18 years or older, who were HIV-negative and previous severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection-free, were eligible to participate and were randomly assigned to receive the vaccine at a dose of 1 x 10(11) viral particles per mL or 5 x 10(10) viral particles per mL, or placebo. Investigators allocated participants at a ratio of 2:1:1 to receive a single injection intramuscularly in the arm. The randomisation list (block size 4) was generated by an independent statistician. Participants, investigators, and staff undertaking laboratory analyses were masked to group allocation. The primary endpoints for immunogenicity were the geometric mean titres (GMTs) of specific ELISA antibody responses to the receptor binding domain (RBD) and neutralising antibody responses at day 28. The primary endpoint for safety evaluation was the incidence of adverse reactions within 14 days. All recruited participants who received at least one dose were included in the primary and safety analyses. This study is registered with ClinicalTrials.gov, NCT04341389. FINDINGS: 603 volunteers were recruited and screened for eligibility between April 11 and 16, 2020. 508 eligible participants (50% male; mean age 39.7 years, SD 12.5) consented to participate in the trial and were randomly assigned to receive the vaccine (1 x 10(11) viral particles n=253; 5 x 10(10) viral particles n=129) or placebo (n=126). In the 1 x 10(11) and 5 x 10(10) viral particles dose groups, the RBD-specific ELISA antibodies peaked at 656.5 (95% CI 575.2-749.2) and 571.0 (467.6-697.3), with seroconversion rates at 96% (95% CI 93-98) and 97% (92-99), respectively, at day 28. Both doses of the vaccine induced significant neutralising antibody responses to live SARS-CoV-2, with GMTs of 19.5 (95% CI 16.8-22.7) and 18.3 (14.4-23.3) in participants receiving 1 x 10(11) and 5 x 10(10) viral particles, respectively. Specific interferon gamma enzyme-linked immunospot assay responses post vaccination were observed in 227 (90%, 95% CI 85-93) of 253 and 113 (88%, 81-92) of 129 participants in the 1 x 10(11) and 5 x 10(10) viral particles dose groups, respectively. Solicited adverse reactions were reported by 183 (72%) of 253 and 96 (74%) of 129 participants in the 1 x 10(11) and 5 x 10(10) viral particles dose groups, respectively. Severe adverse reactions were reported by 24 (9%) participants in the 1 x 10(11) viral particles dose group and one (1%) participant in the 5 x 10(10) viral particles dose group. No serious adverse reactions were documented. INTERPRETATION: The Ad5-vectored COVID-19 vaccine at 5 x 10(10) viral particles is safe, and induced significant immune responses in the majority of recipients after a single immunisation. FUNDING: National Key R&D Programme of China, National Science and Technology Major Project, and CanSino Biologics.
Recombinant adenovirus serotype 26 (Ad26) and Ad35 vaccine vectors bypass immunity to Ad5 and protect nonhuman primates against ebolavirus challenge
DOI:10.1128/JVI.02407-10
URL
PMID:21325402
[Cited within: 1]

The use of adenoviruses (Ad) as vaccine vectors against a variety of pathogens has demonstrated their capacity to elicit strong antibody and cell-mediated immune responses. Adenovirus serotype C vectors, such as Ad serotype 5 (Ad5), expressing Ebolavirus (EBOV) glycoprotein (GP), protect completely after a single inoculation at a dose of 10(10) viral particles. However, the clinical application of a vaccine based on Ad5 vectors may be hampered, since impairment of Ad5 vaccine efficacy has been demonstrated for humans and nonhuman primates with high levels of preexisting immunity to the vector. Ad26 and Ad35 segregate genetically from Ad5 and exhibit lower seroprevalence in humans, making them attractive vaccine vector alternatives. In the series of studies presented, we show that Ad26 and Ad35 vectors generate robust antigen-specific cell-mediated and humoral immune responses against EBOV GP and that Ad5 immune status does not affect the generation of GP-specific immune responses by these vaccines. We demonstrate partial protection against EBOV by a single-shot Ad26 vaccine and complete protection when this vaccine is boosted by Ad35 1 month later. Increases in efficacy are paralleled by substantial increases in T- and B-cell responses to EBOV GP. These results suggest that Ad26 and Ad35 vectors warrant further development as candidate vaccines for EBOV.
A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity
DOI:10.1371/journal.pone.0040385
URL
PMID:22808149
[Cited within: 1]

Recombinant adenoviruses are among the most promising tools for vaccine antigen delivery. Recently, the development of new vectors has focused on serotypes to which the human population is less exposed in order to circumvent pre-existing anti vector immunity. This study describes the derivation of a new vaccine vector based on a chimpanzee adenovirus, Y25, together with a comparative assessment of its potential to elicit transgene product specific immune responses in mice. The vector was constructed in a bacterial artificial chromosome to facilitate genetic manipulation of genomic clones. In order to conduct a fair head-to-head immunological comparison of multiple adenoviral vectors, we optimised a method for accurate determination of infectious titre, since this parameter exhibits profound natural variability and can confound immunogenicity studies when doses are based on viral particle estimation. Cellular immunogenicity of recombinant E1 E3-deleted vector ChAdY25 was comparable to that of other species E derived chimpanzee adenovirus vectors including ChAd63, the first simian adenovirus vector to enter clinical trials in humans. Furthermore, the prevalence of virus neutralizing antibodies (titre >1:200) against ChAdY25 in serum samples collected from two human populations in the UK and Gambia was particularly low compared to published data for other chimpanzee adenoviruses. These findings support the continued development of new chimpanzee adenovirus vectors, including ChAdY25, for clinical use.
Measles-vectored vaccine approaches against viral infections: a focus on Chikungunya
DOI:10.1080/14760584.2019.1562908
URL
PMID:30601074
[Cited within: 2]

INTRODUCTION: The large global burden of viral infections and especially the rapidly spreading vector-borne diseases and other emerging viral diseases show the need for new approaches in vaccine development. Several new vaccine technology platforms have been developed and are under evaluation. Areas covered: This article discusses the measles vector platform technology derived from the safe and highly efficacious measles virus vaccine. The pipeline of measles-vectored vaccine candidates against viral diseases is reviewed. Particular focus is given to the Chikungunya vaccine candidate as the first measles-vectored vaccine that demonstrated safety, immunogenicity, and functionality of the technology in humans even in the presence of pre-existing anti-measles immunity and thus achieved proof of concept for the technology. Expert commentary: Demonstrating no impact of pre-existing anti-measles immunity in humans on the response to the transgene was fundamental for the technology and indicates that the technology is suitable for large-scale immunization in measles pre-immune populations. The proof of concept in humans combined with a large preclinical track record of safety, immunogenicity, and efficacy for a variety of pathogens suggest the measles vector platform as promising plug-and-play vaccine platform technology for rapid development of effective preventive vaccines against viral and other infectious diseases.
Developing mRNA-vaccine technologies
DOI:10.4161/rna.22269
URL
PMID:23064118
[Cited within: 2]

mRNA vaccines combine desirable immunological properties with an outstanding safety profile and the unmet flexibility of genetic vaccines. Based on in situ protein expression, mRNA vaccines are capable of inducing a balanced immune response comprising both cellular and humoral immunity while not subject to MHC haplotype restriction. In addition, mRNA is an intrinsically safe vector as it is a minimal and only transient carrier of information that does not interact with the genome. Because any protein can be expressed from mRNA without the need to adjust the production process, mRNA vaccines also offer maximum flexibility with respect to development. Taken together, mRNA presents a promising vector that may well become the basis of a game-changing vaccine technology platform. Here, we outline the current knowledge regarding different aspects that should be considered when developing an mRNA-based vaccine technology.
mRNA vaccines - a new era in vaccinology
DOI:10.1038/nrd.2017.243
URL
PMID:29326426
[Cited within: 3]

mRNA vaccines represent a promising alternative to conventional vaccine approaches because of their high potency, capacity for rapid development and potential for low-cost manufacture and safe administration. However, their application has until recently been restricted by the instability and inefficient in vivo delivery of mRNA. Recent technological advances have now largely overcome these issues, and multiple mRNA vaccine platforms against infectious diseases and several types of cancer have demonstrated encouraging results in both animal models and humans. This Review provides a detailed overview of mRNA vaccines and considers future directions and challenges in advancing this promising vaccine platform to widespread therapeutic use.
In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies
DOI:10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#
URL
PMID:10602021
[Cited within: 1]

To study the efficiency of RNA-based vaccines, RNA coding for the model antigen beta-galactosidase (beta-gal) was transcribed in vitro from a lacZ gene flanked by stabilizing Xenopus laevis beta-globin 5' and 3' sequences and was protected from RNase degradation by condensation with the polycationic peptide protamine. The liposome-encapsulated condensed RNA-peptide complex, the condensed RNA-peptide complex without liposome or naked, unprotected RNA, was injected into BALB/c (H-2(d)) mice. All preparations led to protein expression in the local tissue, activation of L(d)-restricted specific cytotoxic T lymphocytes (CTL) and production of IgG antibodies reactive against beta-gal. RNA-triggered CTL were as efficient in the lysis of lacZ-transfected target cells as CTL triggered by a lacZ-DNA eukaryotic expression vector. Immunization with RNA transcribed from a cDNA library from the beta-gal-expressing cell line P13.1 again led to beta-gal-specific CTL and IgG induction. Thus, both naked and protected RNA can be used to elicit a specific immune response in vivo, whereby the protected RNA is stable in vitro for a longer period of time. RNA vaccines can be produced in high amounts and have the same major advantages as DNA vaccines but lack the potentially harmful effect of DNA integration into the genome.
Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes
DOI:10.1016/j.jconrel.2015.08.007
URL
PMID:26264835
[Cited within: 1]

In recent years, in vitro transcribed messenger RNA (mRNA) has emerged as a potential therapeutic platform. To fulfill its promise, effective delivery of mRNA to specific cell types and tissues needs to be achieved. Lipid nanoparticles (LNPs) are efficient carriers for short-interfering RNAs and have entered clinical trials. However, little is known about the potential of LNPs to deliver mRNA. Here, we generated mRNA-LNPs by incorporating HPLC purified, 1-methylpseudouridine-containing mRNA comprising codon-optimized firefly luciferase into stable LNPs. Mice were injected with 0.005-0.250mg/kg doses of mRNA-LNPs by 6 different routes and high levels of protein translation could be measured using in vivo imaging. Subcutaneous, intramuscular and intradermal injection of the LNP-encapsulated mRNA translated locally at the site of injection for up to 10days. For several days, high levels of protein production could be achieved in the lung from the intratracheal administration of mRNA. Intravenous and intraperitoneal and to a lesser extent intramuscular and intratracheal deliveries led to trafficking of mRNA-LNPs systemically resulting in active translation of the mRNA in the liver for 1-4 days. Our results demonstrate that LNPs are appropriate carriers for mRNA in vivo and have the potential to become valuable tools for delivering mRNA encoding therapeutic proteins.
An mRNA vaccine against SARS-CoV-2 - preliminary report
DOI:10.1056/NEJMoa2022483
URL
PMID:32663912
[Cited within: 1]

BACKGROUND: The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late 2019 and spread globally, prompting an international effort to accelerate development of a vaccine. The candidate vaccine mRNA-1273 encodes the stabilized prefusion SARS-CoV-2 spike protein. METHODS: We conducted a phase 1, dose-escalation, open-label trial including 45 healthy adults, 18 to 55 years of age, who received two vaccinations, 28 days apart, with mRNA-1273 in a dose of 25 mug, 100 mug, or 250 mug. There were 15 participants in each dose group. RESULTS: After the first vaccination, antibody responses were higher with higher dose (day 29 enzyme-linked immunosorbent assay anti-S-2P antibody geometric mean titer [GMT], 40,227 in the 25-mug group, 109,209 in the 100-mug group, and 213,526 in the 250-mug group). After the second vaccination, the titers increased (day 57 GMT, 299,751, 782,719, and 1,192,154, respectively). After the second vaccination, serum-neutralizing activity was detected by two methods in all participants evaluated, with values generally similar to those in the upper half of the distribution of a panel of control convalescent serum specimens. Solicited adverse events that occurred in more than half the participants included fatigue, chills, headache, myalgia, and pain at the injection site. Systemic adverse events were more common after the second vaccination, particularly with the highest dose, and three participants (21%) in the 250-mug dose group reported one or more severe adverse events. CONCLUSIONS: The mRNA-1273 vaccine induced anti-SARS-CoV-2 immune responses in all participants, and no trial-limiting safety concerns were identified. These findings support further development of this vaccine. (Funded by the National Institute of Allergy and Infectious Diseases and others; mRNA-1273 ClinicalTrials.gov number, NCT04283461).
Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults
DOI:10.1038/s41586-020-2639-4
URL
PMID:32785213
[Cited within: 2]

In March 2020, the World Health Organization (WHO) declared coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)(1), a pandemic. With rapidly accumulating numbers of cases and deaths reported globally(2), a vaccine is urgently needed. Here we report the available safety, tolerability and immunogenicity data from an ongoing placebo-controlled, observer-blinded dose-escalation study (ClinicalTrials.gov identifier NCT04368728) among 45 healthy adults (18-55 years of age), who were randomized to receive 2 doses-separated by 21 days-of 10 mug, 30 mug or 100 mug of BNT162b1. BNT162b1 is a lipid-nanoparticle-formulated, nucleoside-modified mRNA vaccine that encodes the trimerized receptor-binding domain (RBD) of the spike glycoprotein of SARS-CoV-2. Local reactions and systemic events were dose-dependent, generally mild to moderate, and transient. A second vaccination with 100 mug was not administered because of the increased reactogenicity and a lack of meaningfully increased immunogenicity after a single dose compared with the 30-mug dose. RBD-binding IgG concentrations and SARS-CoV-2 neutralizing titres in sera increased with dose level and after a second dose. Geometric mean neutralizing titres reached 1.9-4.6-fold that of a panel of COVID-19 convalescent human sera, which were obtained at least 14 days after a positive SARS-CoV-2 PCR. These results support further evaluation of this mRNA vaccine candidate.
Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability
DOI:10.1038/mt.2008.200
URL
PMID:18797453
[Cited within: 1]

In vitro-transcribed mRNAs encoding physiologically important proteins have considerable potential for therapeutic applications. However, in its present form, mRNA is unfeasible for clinical use because of its labile and immunogenic nature. Here, we investigated whether incorporation of naturally modified nucleotides into transcripts would confer enhanced biological properties to mRNA. We found that mRNAs containing pseudouridines have a higher translational capacity than unmodified mRNAs when tested in mammalian cells and lysates or administered intravenously into mice at 0.015-0.15 mg/kg doses. The delivered mRNA and the encoded protein could be detected in the spleen at 1, 4, and 24 hours after the injection, where both products were at significantly higher levels when pseudouridine-containing mRNA was administered. Even at higher doses, only the unmodified mRNA was immunogenic, inducing high serum levels of interferon-alpha (IFN-alpha). These findings indicate that nucleoside modification is an effective approach to enhance stability and translational capacity of mRNA while diminishing its immunogenicity in vivo. Improved properties conferred by pseudouridine make such mRNA a promising tool for both gene replacement and vaccination.
Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine
DOI:10.1056/NEJMoa2034577
URL
PMID:33301246
[Cited within: 1]

BACKGROUND: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and the resulting coronavirus disease 2019 (Covid-19) have afflicted tens of millions of people in a worldwide pandemic. Safe and effective vaccines are needed urgently. METHODS: In an ongoing multinational, placebo-controlled, observer-blinded, pivotal efficacy trial, we randomly assigned persons 16 years of age or older in a 1:1 ratio to receive two doses, 21 days apart, of either placebo or the BNT162b2 vaccine candidate (30 mug per dose). BNT162b2 is a lipid nanoparticle-formulated, nucleoside-modified RNA vaccine that encodes a prefusion stabilized, membrane-anchored SARS-CoV-2 full-length spike protein. The primary end points were efficacy of the vaccine against laboratory-confirmed Covid-19 and safety. RESULTS: A total of 43,548 participants underwent randomization, of whom 43,448 received injections: 21,720 with BNT162b2 and 21,728 with placebo. There were 8 cases of Covid-19 with onset at least 7 days after the second dose among participants assigned to receive BNT162b2 and 162 cases among those assigned to placebo; BNT162b2 was 95% effective in preventing Covid-19 (95% credible interval, 90.3 to 97.6). Similar vaccine efficacy (generally 90 to 100%) was observed across subgroups defined by age, sex, race, ethnicity, baseline body-mass index, and the presence of coexisting conditions. Among 10 cases of severe Covid-19 with onset after the first dose, 9 occurred in placebo recipients and 1 in a BNT162b2 recipient. The safety profile of BNT162b2 was characterized by short-term, mild-to-moderate pain at the injection site, fatigue, and headache. The incidence of serious adverse events was low and was similar in the vaccine and placebo groups. CONCLUSIONS: A two-dose regimen of BNT162b2 conferred 95% protection against Covid-19 in persons 16 years of age or older. Safety over a median of 2 months was similar to that of other viral vaccines. (Funded by BioNTech and Pfizer; ClinicalTrials.gov number, NCT04368728.).
Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine
DOI:10.1056/NEJMoa2035389
URL
PMID:33378609
[Cited within: 1]

Immunogenicity of a DNA vaccine candidate for COVID-19
DOI:10.1038/s41467-020-16505-0
URL
PMID:32433465
[Cited within: 2]

The coronavirus family member, SARS-CoV-2 has been identified as the causal agent for the pandemic viral pneumonia disease, COVID-19. At this time, no vaccine is available to control further dissemination of the disease. We have previously engineered a synthetic DNA vaccine targeting the MERS coronavirus Spike (S) protein, the major surface antigen of coronaviruses, which is currently in clinical study. Here we build on this prior experience to generate a synthetic DNA-based vaccine candidate targeting SARS-CoV-2 S protein. The engineered construct, INO-4800, results in robust expression of the S protein in vitro. Following immunization of mice and guinea pigs with INO-4800 we measure antigen-specific T cell responses, functional antibodies which neutralize the SARS-CoV-2 infection and block Spike protein binding to the ACE2 receptor, and biodistribution of SARS-CoV-2 targeting antibodies to the lungs. This preliminary dataset identifies INO-4800 as a potential COVID-19 vaccine candidate, supporting further translational study.
DNA vaccines against COVID-19: perspectives and challenges
DOI:10.1016/j.lfs.2020.118919
URL
PMID:33352173
[Cited within: 3]

The coronavirus disease 2019 (COVID-19) is caused by a novel coronavirus known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is associated with several fatal cases worldwide. The rapid spread of this pathogen and the increasing number of cases highlight the urgent development of vaccines. Among the technologies available for vaccine development, DNA vaccination is a promising alternative to conventional vaccines. Since its discovery in the 1990s, it has been of great interest because of its ability to elicit both humoral and cellular immune responses while showing relevant advantages regarding producibility, stability, and storage. This review aimed to summarize the current knowledge and advancements on DNA vaccines against COVID-19, particularly those in clinical trials.
Alum adjuvant boosts adaptive immunity by inducing uric acid and activating inflammatory dendritic cells
URL PMID:18362170 [Cited within: 1]
CpG DNA as a vaccine adjuvant
DOI:10.1586/erv.10.174
URL
PMID:21506647
[Cited within: 2]

Synthetic oligodeoxynucleotides (ODNs) containing unmethylated CpG motifs trigger cells that express Toll-like receptor 9 (including human plasmacytoid dendritic cells and B cells) to mount an innate immune response characterized by the production of Th1 and proinflammatory cytokines. When used as vaccine adjuvants, CpG ODNs improve the function of professional antigen-presenting cells and boost the generation of humoral and cellular vaccine-specific immune responses. These effects are optimized by maintaining ODNs and vaccine in close proximity. The adjuvant properties of CpG ODNs are observed when administered either systemically or mucosally, and persist in immunocompromised hosts. Preclinical studies indicate that CpG ODNs improve the activity of vaccines targeting infectious diseases and cancer. Clinical trials demonstrate that CpG ODNs have a good safety profile and increase the immunogenicity of coadministered vaccines.
The origins of the vaccine cold chain and a glimpse of the future
DOI:10.1016/j.vaccine.2016.11.097
URL
PMID:28364918
[Cited within: 2]

International efforts to eradicate smallpox in the 1960s and 1970s provided the foundation for efforts to expand immunization programmes, including work to develop immunization supply chains. The need to create a reliable system to keep vaccines cold during the lengthy journey from the manufacturer to the point of use, even in remote areas, was a crucial concern during the early days of the Expanded Programme on Immunization. The vaccine cold chain was deliberately separated from other medical distribution systems to assure timely access to and control of vaccines and injection materials. The story of the early development of the vaccine cold chain shows how a number of challenges were overcome with technological and human resource solutions. For example, the lack of methods to monitor exposure of vaccines to heat during transport and storage led to many innovations, including temperature-sensitive vaccine vial monitors and better methods to record and communicate temperatures in vaccine stores. The need for appropriate equipment to store and transport vaccines in tropical developing countries led to innovations in refrigeration equipment as well as the introduction and widespread adoption of novel high performance vaccine cold-boxes and carriers. New technologies also helped to make injection safer. Underlying this work on technologies and equipment was a major effort to develop the human resources required to manage and implement the immunization supply chain. This included creating foundational policies and a management infrastructure; providing training for managers, health workers, technicians, and others. The vaccine cold chain has contributed to one of the world's public health success stories and provides three priority lessons for future: the vaccine supply chain needs to be integrated with other public health supplies, re-designed for efficiency and effectiveness and work is needed in the longer term to eliminate the need for refrigeration in the supply chain.
Storage and Handling of Immunobiologics
https://www.cdc.gov/vaccines/hcp/acip-recs/general-recs/storage.html. Accessed by January 10,
Addressing the cold reality of mRNA vaccine stability
URL PMID:33321139 [Cited within: 4]
thermostable mRNA vaccine against COVID-19
URL PMID:32795413 [Cited within: 1]
Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia
DOI:10.1016/S0140-6736(20)31866-3
URL
PMID:32896291
[Cited within: 1]

BACKGROUND: We developed a heterologous COVID-19 vaccine consisting of two components, a recombinant adenovirus type 26 (rAd26) vector and a recombinant adenovirus type 5 (rAd5) vector, both carrying the gene for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike glycoprotein (rAd26-S and rAd5-S). We aimed to assess the safety and immunogenicity of two formulations (frozen and lyophilised) of this vaccine. METHODS: We did two open, non-randomised phase 1/2 studies at two hospitals in Russia. We enrolled healthy adult volunteers (men and women) aged 18-60 years to both studies. In phase 1 of each study, we administered intramuscularly on day 0 either one dose of rAd26-S or one dose of rAd5-S and assessed the safety of the two components for 28 days. In phase 2 of the study, which began no earlier than 5 days after phase 1 vaccination, we administered intramuscularly a prime-boost vaccination, with rAd26-S given on day 0 and rAd5-S on day 21. Primary outcome measures were antigen-specific humoral immunity (SARS-CoV-2-specific antibodies measured by ELISA on days 0, 14, 21, 28, and 42) and safety (number of participants with adverse events monitored throughout the study). Secondary outcome measures were antigen-specific cellular immunity (T-cell responses and interferon-gamma concentration) and change in neutralising antibodies (detected with a SARS-CoV-2 neutralisation assay). These trials are registered with ClinicalTrials.gov, NCT04436471 and NCT04437875. FINDINGS: Between June 18 and Aug 3, 2020, we enrolled 76 participants to the two studies (38 in each study). In each study, nine volunteers received rAd26-S in phase 1, nine received rAd5-S in phase 1, and 20 received rAd26-S and rAd5-S in phase 2. Both vaccine formulations were safe and well tolerated. The most common adverse events were pain at injection site (44 [58%]), hyperthermia (38 [50%]), headache (32 [42%]), asthenia (21 [28%]), and muscle and joint pain (18 [24%]). Most adverse events were mild and no serious adverse events were detected. All participants produced antibodies to SARS-CoV-2 glycoprotein. At day 42, receptor binding domain-specific IgG titres were 14 703 with the frozen formulation and 11 143 with the lyophilised formulation, and neutralising antibodies were 49.25 with the frozen formulation and 45.95 with the lyophilised formulation, with a seroconversion rate of 100%. Cell-mediated responses were detected in all participants at day 28, with median cell proliferation of 2.5% CD4(+) and 1.3% CD8(+) with the frozen formulation, and a median cell proliferation of 1.3% CD4(+) and 1.1% CD8(+) with the lyophilised formulation. INTERPRETATION: The heterologous rAd26 and rAd5 vector-based COVID-19 vaccine has a good safety profile and induced strong humoral and cellular immune responses in participants. Further investigation is needed of the effectiveness of this vaccine for prevention of COVID-19. FUNDING: Ministry of Health of the Russian Federation.
mRNA vaccine: a potential therapeutic strategy
DOI:10.1186/s12943-021-01311-z
URL
PMID:33593376
[Cited within: 1]

mRNA vaccines have tremendous potential to fight against cancer and viral diseases due to superiorities in safety, efficacy and industrial production. In recent decades, we have witnessed the development of different kinds of mRNAs by sequence optimization to overcome the disadvantage of excessive mRNA immunogenicity, instability and inefficiency. Based on the immunological study, mRNA vaccines are coupled with immunologic adjuvant and various delivery strategies. Except for sequence optimization, the assistance of mRNA-delivering strategies is another method to stabilize mRNAs and improve their efficacy. The understanding of increasing the antigen reactiveness gains insight into mRNA-induced innate immunity and adaptive immunity without antibody-dependent enhancement activity. Therefore, to address the problem, scientists further exploited carrier-based mRNA vaccines (lipid-based delivery, polymer-based delivery, peptide-based delivery, virus-like replicon particle and cationic nanoemulsion), naked mRNA vaccines and dendritic cells-based mRNA vaccines. The article will discuss the molecular biology of mRNA vaccines and underlying anti-virus and anti-tumor mechanisms, with an introduction of their immunological phenomena, delivery strategies, their importance on Corona Virus Disease 2019 (COVID-19) and related clinical trials against cancer and viral diseases. Finally, we will discuss the challenge of mRNA vaccines against bacterial and parasitic diseases.
The influence of antigen organization on B cell responsiveness
DOI:10.1126/science.8248784
URL
PMID:8248784
[Cited within: 1]

The influence of antigen epitope density and order on B cell induction and antibody production was assessed with the glycoprotein of vesicular stomatitis virus serotype Indiana [VSV-G (IND)]. VSV-G (IND) can be found in a highly repetitive form the envelope of VSV-IND and in a poorly organized form on the surface of infected cells. In VSV-G (IND) transgenic mice, B cells were unresponsive to the poorly organized VSV-G (IND) present as self antigen but responded promptly to the same antigen presented in the highly organized form. Thus, antigen organization influences B cell tolerance.
Enhancing antibody response against small molecular hapten with tobacco mosaic virus as a polyvalent carrier
DOI:10.1002/cbic.201500028
URL
PMID:25914312
[Cited within: 1]

Virus nanoparticles (VNPs) have been applied as carrier proteins for effective vaccine development. In this paper, we report the usage of tobacco mosaic virus (TMV) as a carrier for the display of the small molecule estriol (E3), a weakly immunogenic hapten. A highly efficient copper (I)-catalyzed azide-alkyne cycloaddition reaction (CuAAC) was performed for the conjugation of E3 onto TMV capsid at tyrosine (Tyr) 139, by which the antigen density could be controlled. The immune properties of these constructs were evaluated in mice. We found that a strong and long-term antibody response was elicited by conjugating a high density of small molecular haptens on TMV through an oligo(ethylene glycol) (OEG) linker, likely due to the effective activation of B-cells. This study suggests that TMV can serve as a promising platform to induce strong humoral immune responses and that the optimized conjugation strategy was critical to produce high quality antibodies.
Polymer-protein core-shell nanoparticles for enhanced antigen immunogenicity
Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors
Public Now Divided Over Whether To Get COVID-19 Vaccine
https://www.pewresearch.org/science/2020/09/17/u-s-public-now-divided-over-whether-to-get-covid-19-vaccine/. Accessed by January 10,
Covid-19: New coronavirus variant is identified in UK
DOI:10.1136/bmj.m4857 URL PMID:33328153 [Cited within: 1]
Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein
DOI:10.1038/d41586-020-00940-6 URL PMID:33772232 [Cited within: 1]
COVID-19 vaccine development and a potential nanomaterial path forward
DOI:10.1038/s41565-020-0737-y
URL
PMID:32669664
[Cited within: 1]

The COVID-19 pandemic has infected millions of people with no clear signs of abatement owing to the high prevalence, long incubation period and lack of established treatments or vaccines. Vaccines are the most promising solution to mitigate new viral strains. The genome sequence and protein structure of the 2019-novel coronavirus (nCoV or SARS-CoV-2) were made available in record time, allowing the development of inactivated or attenuated viral vaccines along with subunit vaccines for prophylaxis and treatment. Nanotechnology benefits modern vaccine design since nanomaterials are ideal for antigen delivery, as adjuvants, and as mimics of viral structures. In fact, the first vaccine candidate launched into clinical trials is an mRNA vaccine delivered via lipid nanoparticles. To eradicate pandemics, present and future, a successful vaccine platform must enable rapid discovery, scalable manufacturing and global distribution. Here, we review current approaches to COVID-19 vaccine development and highlight the role of nanotechnology and advanced manufacturing.